Probe CUST_25316_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25316_PI426222305 | JHI_St_60k_v1 | DMT400034702 | AAGAGGAAAGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAG |
All Microarray Probes Designed to Gene DMG402013333
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25480_PI426222305 | JHI_St_60k_v1 | DMT400034701 | TTTATCTTGATGAAAAGAACCGCCCTCTGTTAATTCACTGCAAAAGAGGAAAGATGAACG |
| CUST_25459_PI426222305 | JHI_St_60k_v1 | DMT400034700 | TGGTTGTTGCTGTTAAAATCCCAACCTTTCTACTCGAAGAGTTGTGAGTTTGTTAAAAAA |
| CUST_25316_PI426222305 | JHI_St_60k_v1 | DMT400034702 | AAGAGGAAAGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAG |
| CUST_25464_PI426222305 | JHI_St_60k_v1 | DMT400034703 | TTTATCTTGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAGG |
Microarray Signals from CUST_25316_PI426222305
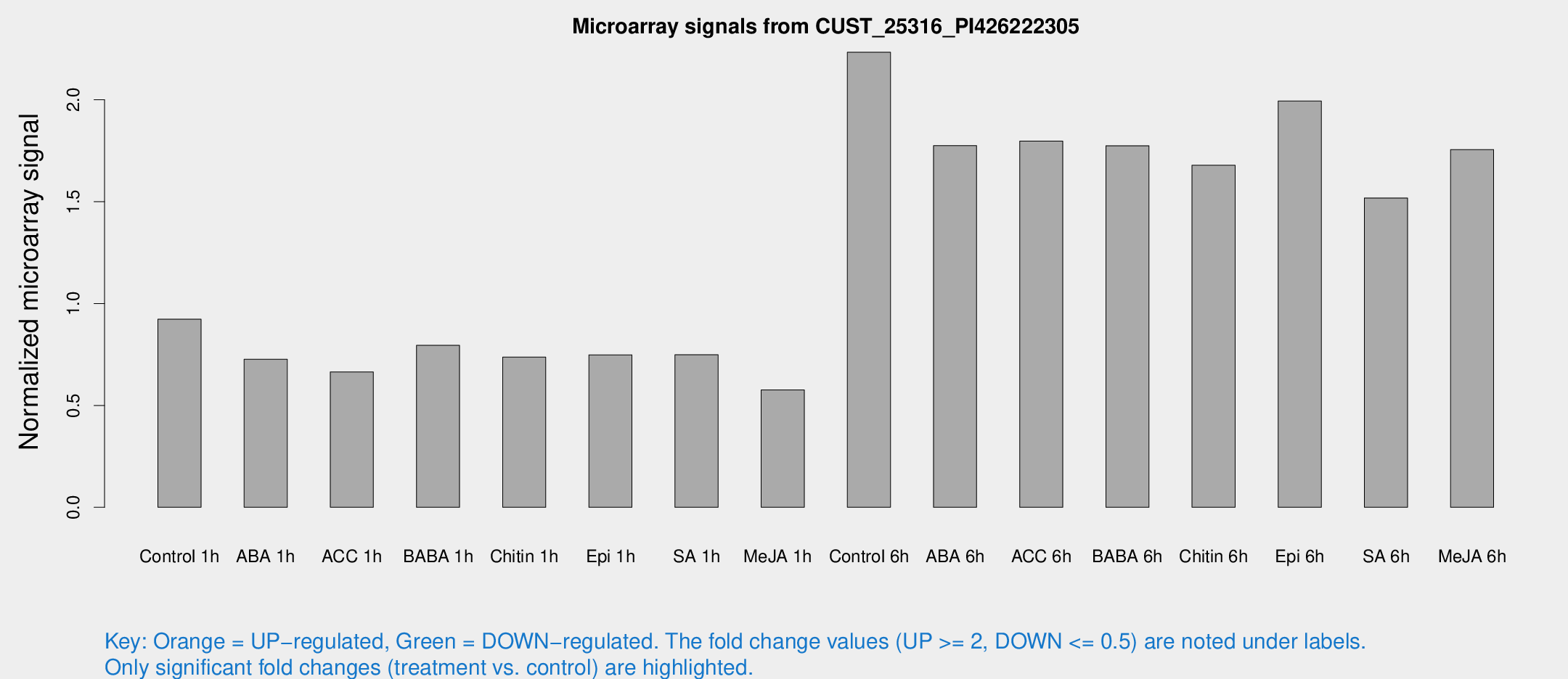
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 690.101 | 79.7891 | 0.923161 | 0.053489 |
| ABA 1h | 474.623 | 27.5882 | 0.726801 | 0.0599755 |
| ACC 1h | 522.536 | 92.2601 | 0.664596 | 0.0796579 |
| BABA 1h | 587.317 | 110.83 | 0.794848 | 0.0861515 |
| Chitin 1h | 490.534 | 28.5822 | 0.7373 | 0.0428528 |
| Epi 1h | 481.108 | 37.8292 | 0.748218 | 0.0479838 |
| SA 1h | 575.532 | 65.6519 | 0.748669 | 0.0442244 |
| Me-JA 1h | 347.828 | 20.4065 | 0.576086 | 0.041717 |
| Control 6h | 1899.91 | 714.387 | 2.2334 | 0.75351 |
| ABA 6h | 1454.41 | 275.693 | 1.77526 | 0.310925 |
| ACC 6h | 1555.28 | 238.069 | 1.79711 | 0.103864 |
| BABA 6h | 1489.27 | 190.916 | 1.77454 | 0.200645 |
| Chitin 6h | 1321.06 | 76.5033 | 1.6785 | 0.0970327 |
| Epi 6h | 1689.41 | 228.57 | 1.99366 | 0.230142 |
| SA 6h | 1259.6 | 409.517 | 1.51823 | 0.421421 |
| Me-JA 6h | 1320.75 | 191.065 | 1.75577 | 0.130361 |
Source Transcript PGSC0003DMT400034702 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05000.2 | +1 | 1e-74 | 241 | 122/195 (63%) | Phosphotyrosine protein phosphatases superfamily protein | chr1:1425660-1428393 FORWARD LENGTH=247 |
