Probe CUST_25464_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25464_PI426222305 | JHI_St_60k_v1 | DMT400034703 | TTTATCTTGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAGG |
All Microarray Probes Designed to Gene DMG402013333
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25480_PI426222305 | JHI_St_60k_v1 | DMT400034701 | TTTATCTTGATGAAAAGAACCGCCCTCTGTTAATTCACTGCAAAAGAGGAAAGATGAACG |
| CUST_25459_PI426222305 | JHI_St_60k_v1 | DMT400034700 | TGGTTGTTGCTGTTAAAATCCCAACCTTTCTACTCGAAGAGTTGTGAGTTTGTTAAAAAA |
| CUST_25316_PI426222305 | JHI_St_60k_v1 | DMT400034702 | AAGAGGAAAGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAG |
| CUST_25464_PI426222305 | JHI_St_60k_v1 | DMT400034703 | TTTATCTTGGAGCCACCATTGGTCAACATCCCAGAGGAAACAATAAAGGAAGCTTTGAGG |
Microarray Signals from CUST_25464_PI426222305
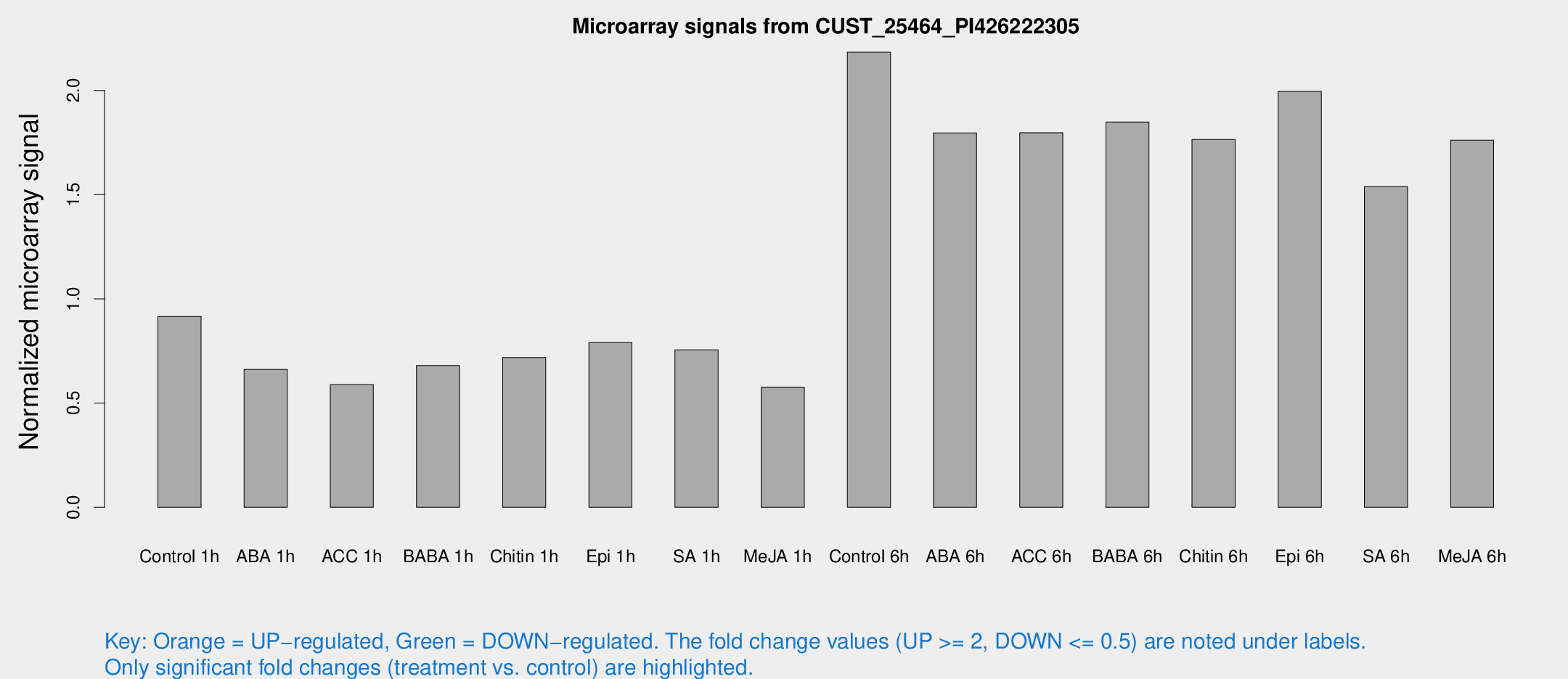
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 584.232 | 53.3214 | 0.915846 | 0.0531176 |
| ABA 1h | 370.203 | 21.6058 | 0.661286 | 0.0385755 |
| ACC 1h | 424.16 | 116.374 | 0.588776 | 0.16525 |
| BABA 1h | 438.897 | 96.1217 | 0.680796 | 0.0992898 |
| Chitin 1h | 410.765 | 28.8135 | 0.71853 | 0.0418686 |
| Epi 1h | 434.915 | 31.0293 | 0.790315 | 0.0460168 |
| SA 1h | 499.818 | 65.6961 | 0.755567 | 0.0547992 |
| Me-JA 1h | 298.214 | 17.5819 | 0.575579 | 0.0338352 |
| Control 6h | 1560.98 | 548.127 | 2.18319 | 0.667944 |
| ABA 6h | 1254.65 | 225.151 | 1.79578 | 0.289824 |
| ACC 6h | 1344.88 | 241.492 | 1.79701 | 0.103891 |
| BABA 6h | 1332.82 | 184.28 | 1.84831 | 0.225373 |
| Chitin 6h | 1191.18 | 69.0595 | 1.76428 | 0.102009 |
| Epi 6h | 1436.98 | 143.109 | 1.99498 | 0.17414 |
| SA 6h | 1055.19 | 284.361 | 1.53875 | 0.321614 |
| Me-JA 6h | 1128.22 | 139.023 | 1.76112 | 0.10818 |
Source Transcript PGSC0003DMT400034703 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G05000.2 | +1 | 1e-67 | 223 | 122/230 (53%) | Phosphotyrosine protein phosphatases superfamily protein | chr1:1425660-1428393 FORWARD LENGTH=247 |
