Probe CUST_14729_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14729_PI426222305 | JHI_St_60k_v1 | DMT400066419 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
All Microarray Probes Designed to Gene DMG401025828
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14484_PI426222305 | JHI_St_60k_v1 | DMT400066422 | CCCCATTTAAAAGCAGCAGCAATTATGTTCAAAAGATAGGTCTTCTATTCTGAGTTTTCT |
| CUST_14615_PI426222305 | JHI_St_60k_v1 | DMT400066420 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14555_PI426222305 | JHI_St_60k_v1 | DMT400066421 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14453_PI426222305 | JHI_St_60k_v1 | DMT400066425 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14729_PI426222305 | JHI_St_60k_v1 | DMT400066419 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
Microarray Signals from CUST_14729_PI426222305
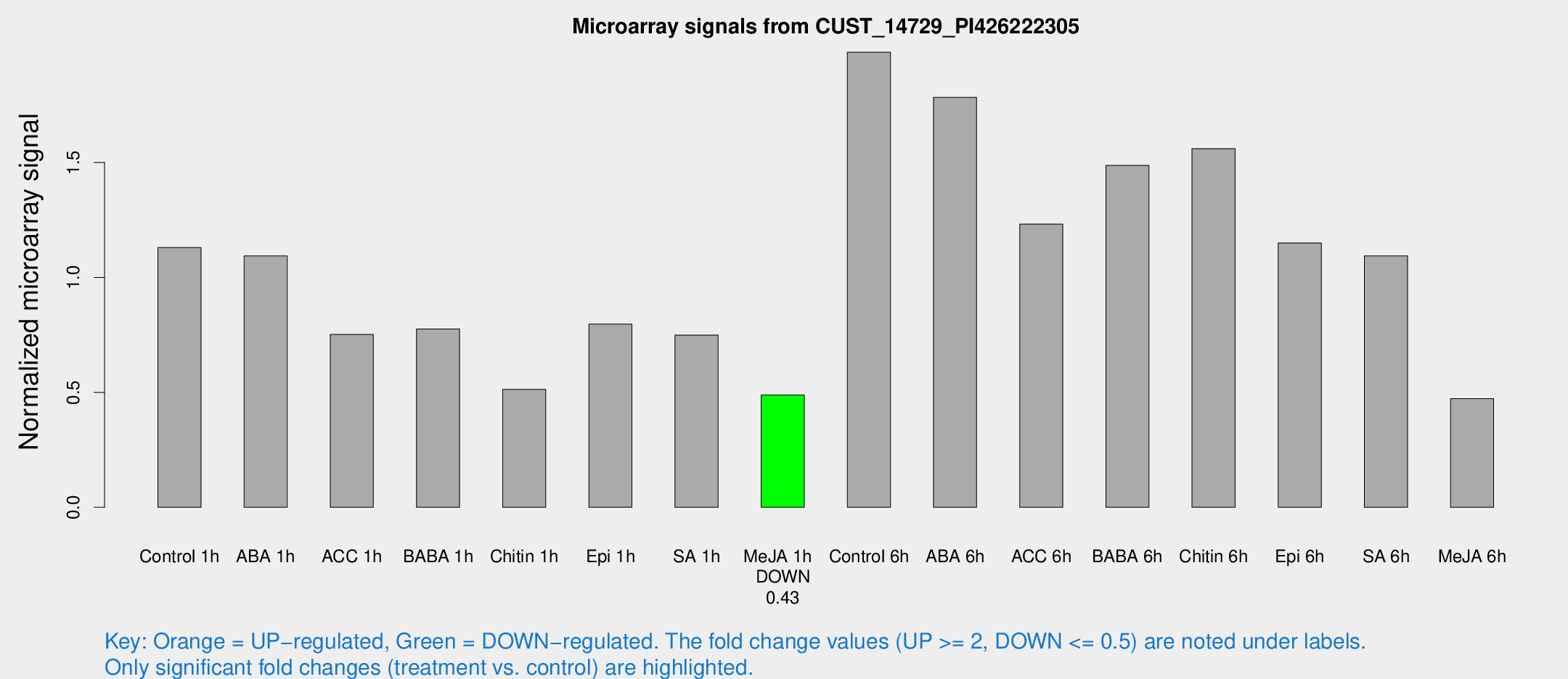
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 56.0472 | 6.83886 | 1.12997 | 0.0878079 |
| ABA 1h | 48.0173 | 6.02903 | 1.09381 | 0.123214 |
| ACC 1h | 42.0728 | 11.9573 | 0.752225 | 0.220955 |
| BABA 1h | 38.1292 | 7.38892 | 0.776107 | 0.0894947 |
| Chitin 1h | 23.2681 | 3.88687 | 0.513337 | 0.134025 |
| Epi 1h | 34.105 | 3.73641 | 0.797106 | 0.0817188 |
| SA 1h | 38.625 | 6.35353 | 0.74938 | 0.153823 |
| Me-JA 1h | 19.7799 | 3.11986 | 0.488765 | 0.07812 |
| Control 6h | 99.0029 | 14.0099 | 1.98009 | 0.156746 |
| ABA 6h | 96.836 | 19.1434 | 1.78383 | 0.301501 |
| ACC 6h | 69.309 | 5.36743 | 1.23227 | 0.136493 |
| BABA 6h | 85.7556 | 18.3956 | 1.48734 | 0.316656 |
| Chitin 6h | 84.5673 | 15.9208 | 1.56051 | 0.351859 |
| Epi 6h | 64.7802 | 10.0965 | 1.15005 | 0.197463 |
| SA 6h | 57.8603 | 18.1506 | 1.09366 | 0.515932 |
| Me-JA 6h | 36.2003 | 18.4224 | 0.472686 | 0.492333 |
Source Transcript PGSC0003DMT400066419 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65940.1 | +2 | 9e-90 | 276 | 136/183 (74%) | beta-hydroxyisobutyryl-CoA hydrolase 1 | chr5:26376830-26379161 REVERSE LENGTH=378 |
