Probe CUST_14484_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14484_PI426222305 | JHI_St_60k_v1 | DMT400066422 | CCCCATTTAAAAGCAGCAGCAATTATGTTCAAAAGATAGGTCTTCTATTCTGAGTTTTCT |
All Microarray Probes Designed to Gene DMG401025828
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14484_PI426222305 | JHI_St_60k_v1 | DMT400066422 | CCCCATTTAAAAGCAGCAGCAATTATGTTCAAAAGATAGGTCTTCTATTCTGAGTTTTCT |
| CUST_14615_PI426222305 | JHI_St_60k_v1 | DMT400066420 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14555_PI426222305 | JHI_St_60k_v1 | DMT400066421 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14453_PI426222305 | JHI_St_60k_v1 | DMT400066425 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14729_PI426222305 | JHI_St_60k_v1 | DMT400066419 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
Microarray Signals from CUST_14484_PI426222305
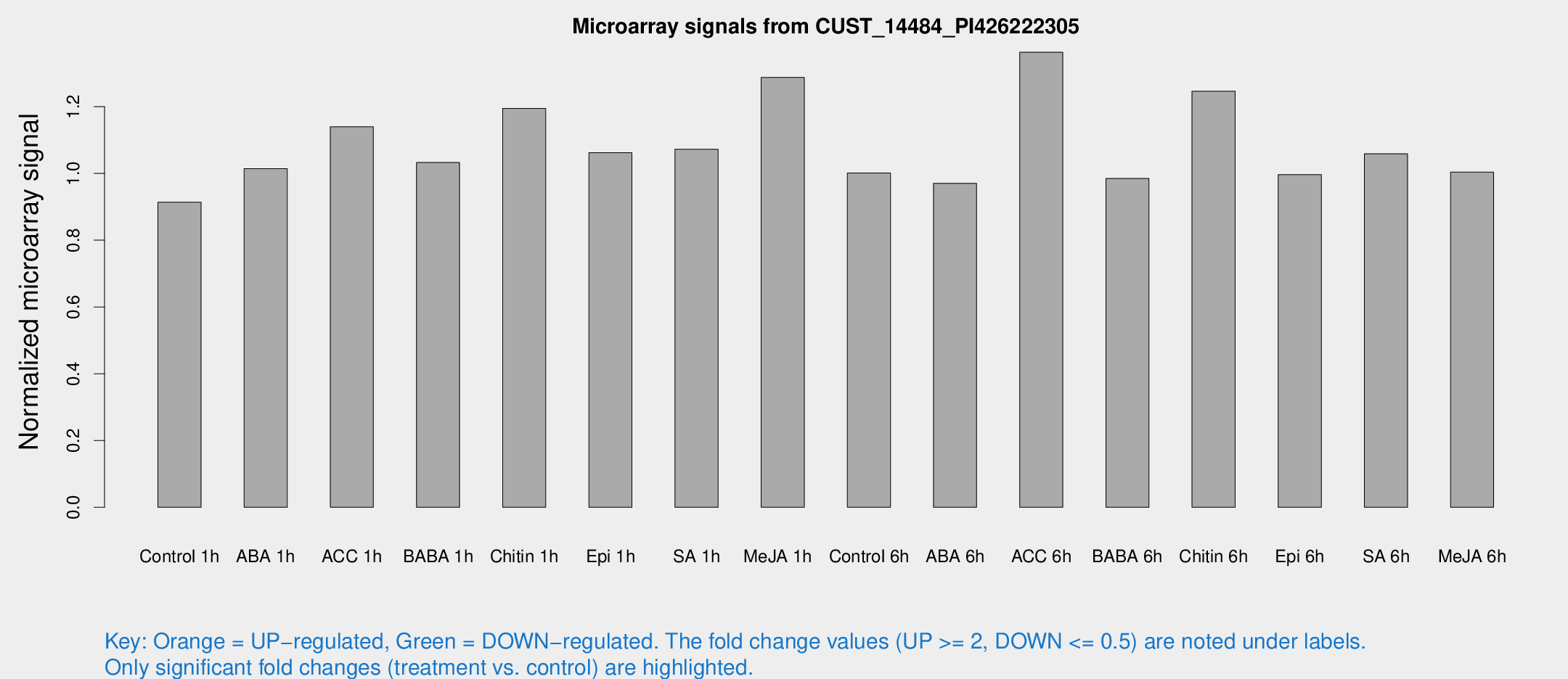
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.0635 | 3.51431 | 0.913685 | 0.529461 |
| ABA 1h | 5.95854 | 3.45297 | 1.01445 | 0.587459 |
| ACC 1h | 7.8793 | 3.91513 | 1.13957 | 0.586174 |
| BABA 1h | 6.60974 | 3.83386 | 1.03245 | 0.598293 |
| Chitin 1h | 7.3074 | 3.66381 | 1.19466 | 0.620751 |
| Epi 1h | 6.11096 | 3.54623 | 1.06183 | 0.614887 |
| SA 1h | 7.58915 | 3.45204 | 1.07217 | 0.529966 |
| Me-JA 1h | 7.03508 | 3.68566 | 1.28743 | 0.68296 |
| Control 6h | 6.66144 | 3.86228 | 1.00099 | 0.579948 |
| ABA 6h | 6.85357 | 3.97721 | 0.970457 | 0.562637 |
| ACC 6h | 11.204 | 4.65113 | 1.3629 | 0.646489 |
| BABA 6h | 7.34215 | 4.27267 | 0.984788 | 0.570752 |
| Chitin 6h | 9.20409 | 4.23215 | 1.24631 | 0.622604 |
| Epi 6h | 7.53055 | 4.40499 | 0.996511 | 0.577035 |
| SA 6h | 6.94887 | 4.02585 | 1.05867 | 0.6133 |
| Me-JA 6h | 6.63889 | 3.85022 | 1.00363 | 0.581448 |
Source Transcript PGSC0003DMT400066422 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65940.1 | +3 | 3e-15 | 79 | 42/90 (47%) | beta-hydroxyisobutyryl-CoA hydrolase 1 | chr5:26376830-26379161 REVERSE LENGTH=378 |
