Probe CUST_14615_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14615_PI426222305 | JHI_St_60k_v1 | DMT400066420 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
All Microarray Probes Designed to Gene DMG401025828
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14484_PI426222305 | JHI_St_60k_v1 | DMT400066422 | CCCCATTTAAAAGCAGCAGCAATTATGTTCAAAAGATAGGTCTTCTATTCTGAGTTTTCT |
| CUST_14615_PI426222305 | JHI_St_60k_v1 | DMT400066420 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14555_PI426222305 | JHI_St_60k_v1 | DMT400066421 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14453_PI426222305 | JHI_St_60k_v1 | DMT400066425 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14729_PI426222305 | JHI_St_60k_v1 | DMT400066419 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
Microarray Signals from CUST_14615_PI426222305
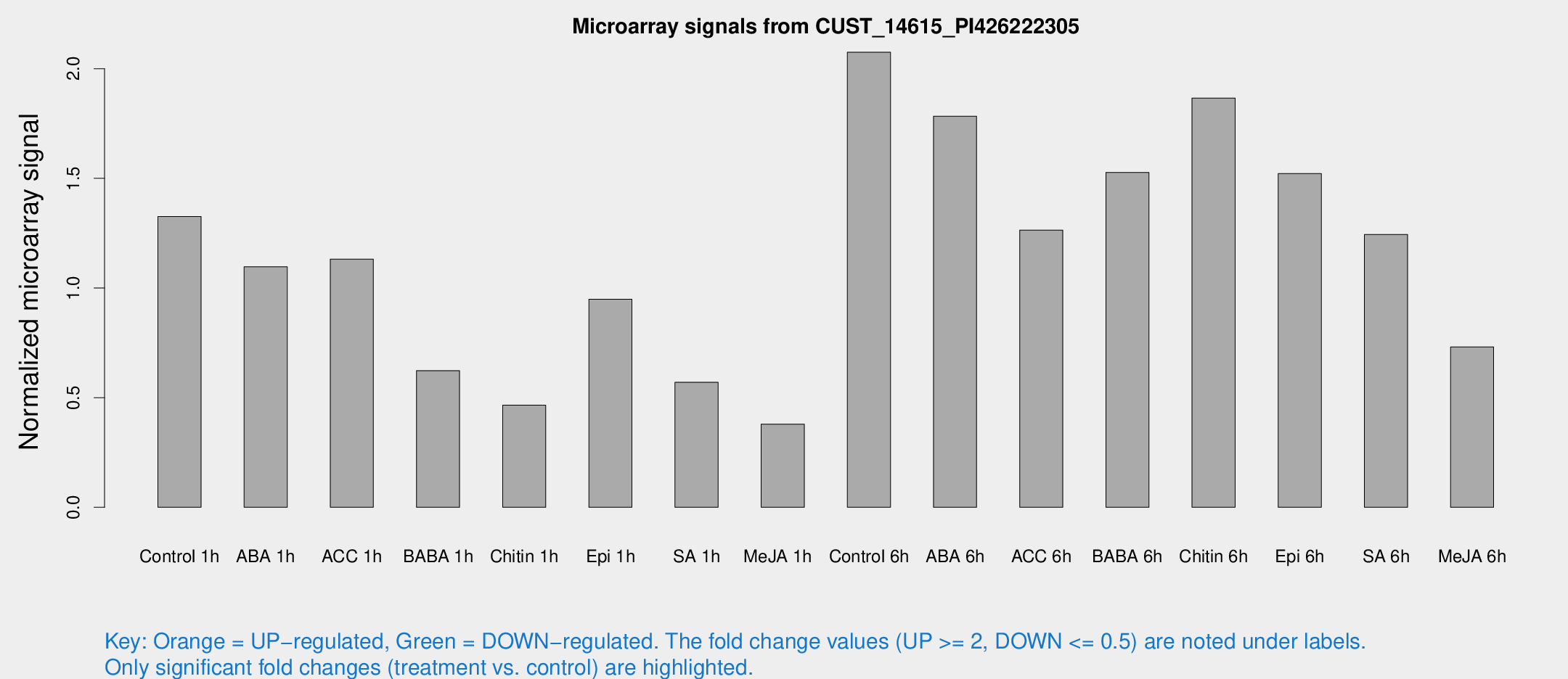
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 47.5541 | 5.29022 | 1.32553 | 0.219191 |
| ABA 1h | 37.8195 | 11.9379 | 1.09703 | 0.343326 |
| ACC 1h | 43.5581 | 9.27322 | 1.13195 | 0.195938 |
| BABA 1h | 25.1649 | 8.26977 | 0.623372 | 0.235593 |
| Chitin 1h | 17.4979 | 6.46333 | 0.465853 | 0.267955 |
| Epi 1h | 30.7193 | 7.24844 | 0.948665 | 0.207268 |
| SA 1h | 24.5801 | 8.20162 | 0.570165 | 0.35204 |
| Me-JA 1h | 12.1196 | 3.62478 | 0.379435 | 0.154344 |
| Control 6h | 75.7074 | 12.6755 | 2.07515 | 0.253011 |
| ABA 6h | 73.2651 | 21.4343 | 1.78354 | 0.447058 |
| ACC 6h | 54.7643 | 12.3761 | 1.26405 | 0.290941 |
| BABA 6h | 63.7023 | 13.1744 | 1.5269 | 0.31476 |
| Chitin 6h | 75.1025 | 17.4604 | 1.86627 | 0.511136 |
| Epi 6h | 80.3949 | 42.043 | 1.52221 | 1.06505 |
| SA 6h | 50.4854 | 20.1783 | 1.24411 | 0.737758 |
| Me-JA 6h | 38.006 | 21.7778 | 0.731433 | 0.531993 |
Source Transcript PGSC0003DMT400066420 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65940.1 | +2 | 5e-49 | 176 | 90/140 (64%) | beta-hydroxyisobutyryl-CoA hydrolase 1 | chr5:26376830-26379161 REVERSE LENGTH=378 |
