Probe CUST_14555_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14555_PI426222305 | JHI_St_60k_v1 | DMT400066421 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
All Microarray Probes Designed to Gene DMG401025828
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14484_PI426222305 | JHI_St_60k_v1 | DMT400066422 | CCCCATTTAAAAGCAGCAGCAATTATGTTCAAAAGATAGGTCTTCTATTCTGAGTTTTCT |
| CUST_14615_PI426222305 | JHI_St_60k_v1 | DMT400066420 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14555_PI426222305 | JHI_St_60k_v1 | DMT400066421 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14453_PI426222305 | JHI_St_60k_v1 | DMT400066425 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14729_PI426222305 | JHI_St_60k_v1 | DMT400066419 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
Microarray Signals from CUST_14555_PI426222305
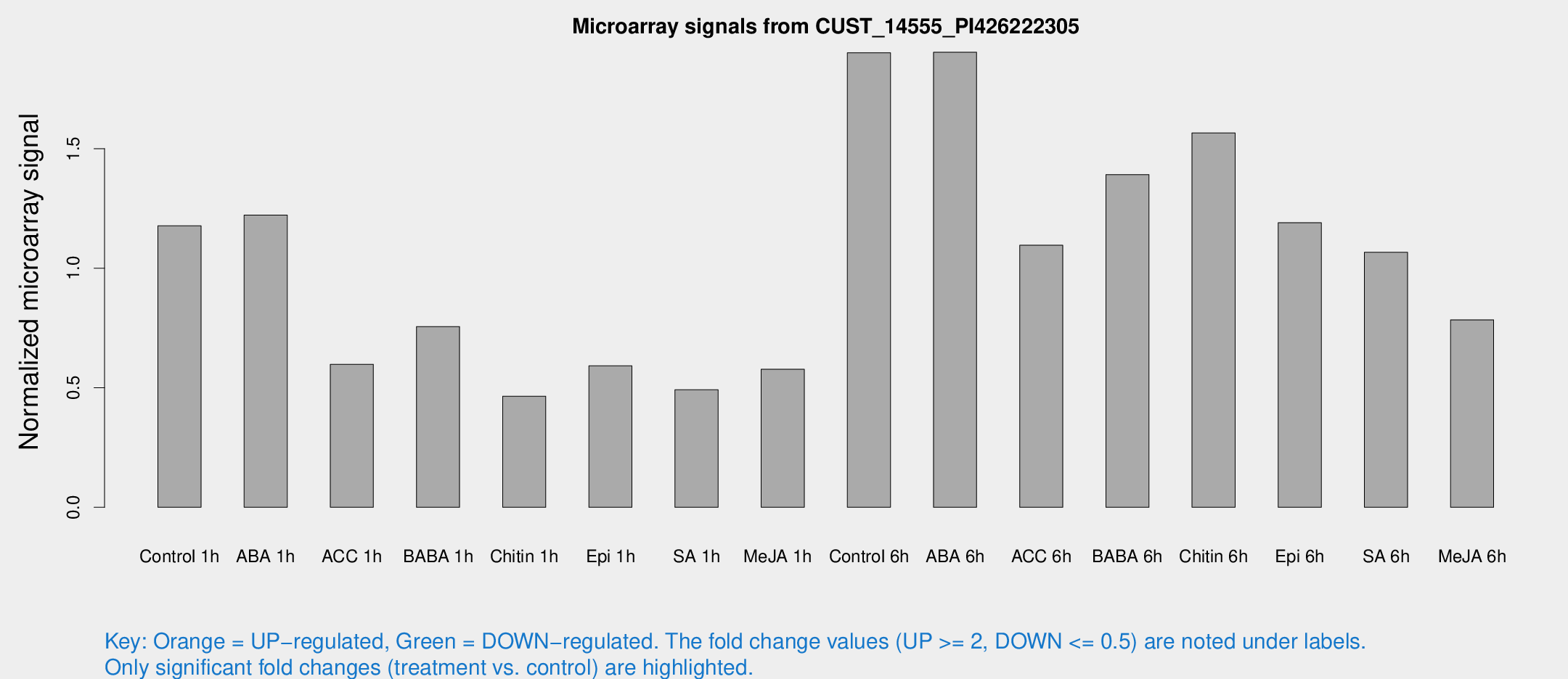
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 51.3892 | 6.41665 | 1.17705 | 0.113169 |
| ABA 1h | 48.2306 | 9.32723 | 1.22199 | 0.292314 |
| ACC 1h | 35.3682 | 13.884 | 0.598139 | 0.412069 |
| BABA 1h | 38.1389 | 13.2443 | 0.755931 | 0.328991 |
| Chitin 1h | 19.4631 | 5.73918 | 0.464591 | 0.13385 |
| Epi 1h | 24.2435 | 7.31683 | 0.59151 | 0.182939 |
| SA 1h | 23.9952 | 6.4816 | 0.491726 | 0.184535 |
| Me-JA 1h | 21.2864 | 4.63433 | 0.577103 | 0.115008 |
| Control 6h | 88.4631 | 25.4806 | 1.90121 | 0.459731 |
| ABA 6h | 89.6748 | 14.4877 | 1.90319 | 0.251029 |
| ACC 6h | 56.3676 | 10.9098 | 1.09615 | 0.23573 |
| BABA 6h | 72.3473 | 18.07 | 1.39148 | 0.375729 |
| Chitin 6h | 75.1193 | 15.8985 | 1.56583 | 0.329436 |
| Epi 6h | 60.179 | 12.1542 | 1.1902 | 0.207828 |
| SA 6h | 48.0633 | 12.0918 | 1.06626 | 0.415654 |
| Me-JA 6h | 37.2258 | 12.5778 | 0.783964 | 0.199147 |
Source Transcript PGSC0003DMT400066421 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65940.1 | +2 | 7e-24 | 104 | 50/56 (89%) | beta-hydroxyisobutyryl-CoA hydrolase 1 | chr5:26376830-26379161 REVERSE LENGTH=378 |
