Probe CUST_11337_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11337_PI426222305 | JHI_St_60k_v1 | DMT400007384 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
All Microarray Probes Designed to Gene DMG400002837
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11054_PI426222305 | JHI_St_60k_v1 | DMT400007385 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11283_PI426222305 | JHI_St_60k_v1 | DMT400007381 | AATAGCTCTTTTTCATTTTGGAGAACAAAATGCAGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11337_PI426222305 | JHI_St_60k_v1 | DMT400007384 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11004_PI426222305 | JHI_St_60k_v1 | DMT400007382 | CTCTGTCTGATCTGGTATGTTTGGAACACCTAATGATTTCTTTAATGAGGAACTCCTCGT |
Microarray Signals from CUST_11337_PI426222305
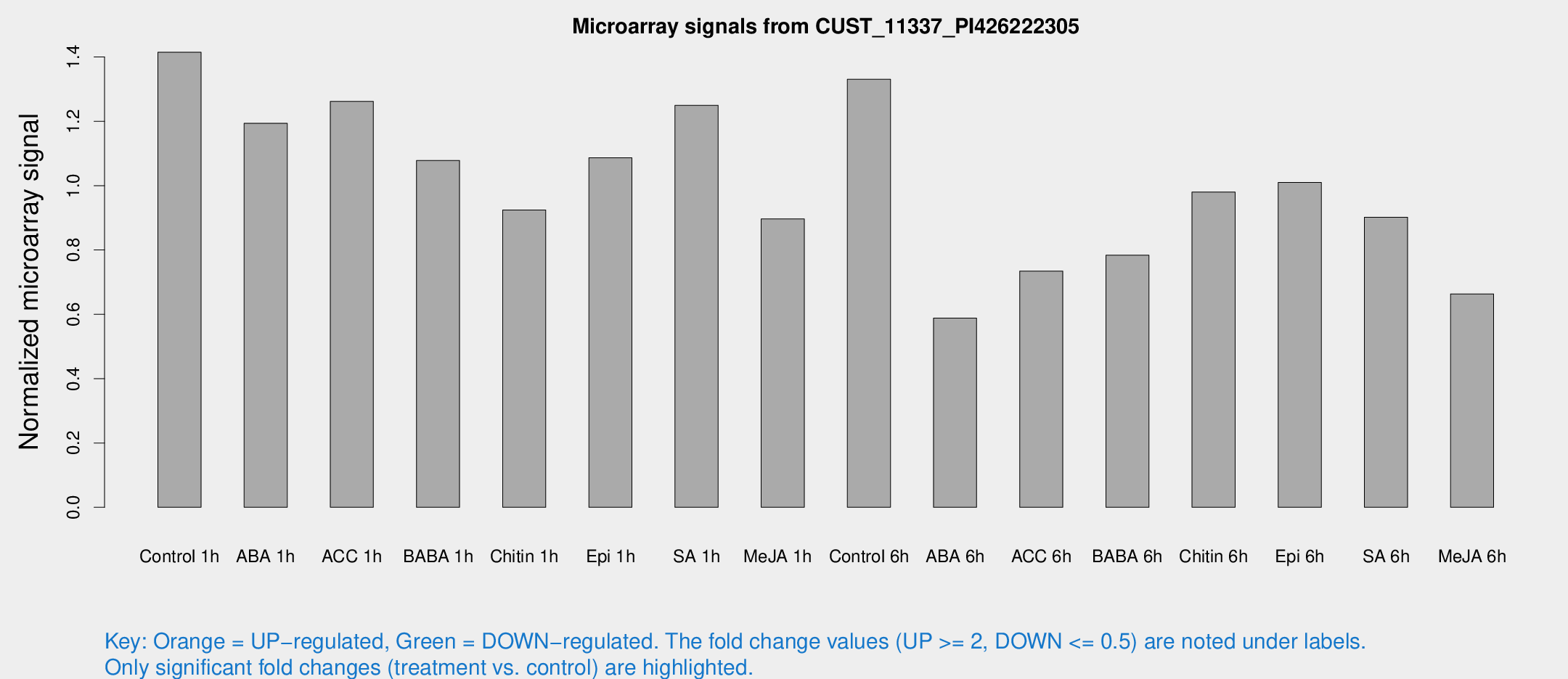
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 354.478 | 63.0282 | 1.41488 | 0.152204 |
| ABA 1h | 259.788 | 27.2201 | 1.19398 | 0.137037 |
| ACC 1h | 326.928 | 59.6843 | 1.26184 | 0.152474 |
| BABA 1h | 259.299 | 39.0901 | 1.07828 | 0.0644725 |
| Chitin 1h | 206.359 | 28.2432 | 0.924441 | 0.0770293 |
| Epi 1h | 229.705 | 14.9856 | 1.08685 | 0.0646189 |
| SA 1h | 314.002 | 24.2519 | 1.24992 | 0.0892038 |
| Me-JA 1h | 178.372 | 10.825 | 0.89664 | 0.0542904 |
| Control 6h | 346.421 | 84.4811 | 1.33078 | 0.255808 |
| ABA 6h | 152.364 | 9.47009 | 0.588071 | 0.0364812 |
| ACC 6h | 210.33 | 34.2995 | 0.734496 | 0.0446498 |
| BABA 6h | 226.154 | 57.5197 | 0.783774 | 0.188319 |
| Chitin 6h | 255.779 | 22.6991 | 0.980322 | 0.0603173 |
| Epi 6h | 280.569 | 31.2027 | 1.00998 | 0.185984 |
| SA 6h | 225.932 | 46.7047 | 0.901813 | 0.118913 |
| Me-JA 6h | 165.954 | 28.8615 | 0.663485 | 0.0660256 |
Source Transcript PGSC0003DMT400007384 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G33290.1 | +3 | 5e-166 | 481 | 238/330 (72%) | P-loop containing nucleoside triphosphate hydrolases superfamily protein | chr1:12074125-12075989 FORWARD LENGTH=379 |
