Probe CUST_11283_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11283_PI426222305 | JHI_St_60k_v1 | DMT400007381 | AATAGCTCTTTTTCATTTTGGAGAACAAAATGCAGACTGAAAGAAGTGTCGATGCTCTGC |
All Microarray Probes Designed to Gene DMG400002837
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11054_PI426222305 | JHI_St_60k_v1 | DMT400007385 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11283_PI426222305 | JHI_St_60k_v1 | DMT400007381 | AATAGCTCTTTTTCATTTTGGAGAACAAAATGCAGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11337_PI426222305 | JHI_St_60k_v1 | DMT400007384 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11004_PI426222305 | JHI_St_60k_v1 | DMT400007382 | CTCTGTCTGATCTGGTATGTTTGGAACACCTAATGATTTCTTTAATGAGGAACTCCTCGT |
Microarray Signals from CUST_11283_PI426222305
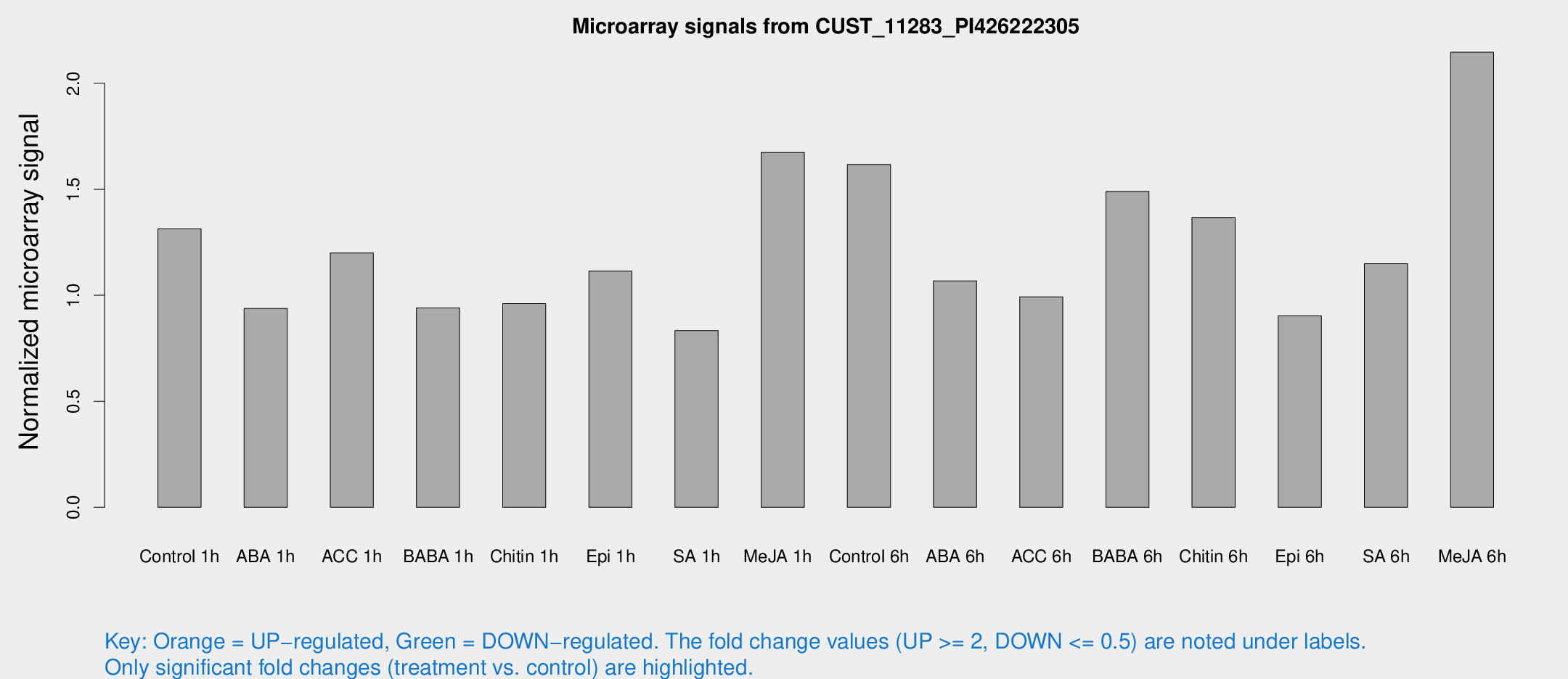
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 10.4104 | 4.75254 | 1.31365 | 0.604168 |
| ABA 1h | 5.42551 | 3.15574 | 0.937536 | 0.542421 |
| ACC 1h | 8.3469 | 3.56942 | 1.19991 | 0.54613 |
| BABA 1h | 5.94098 | 3.44779 | 0.94075 | 0.545854 |
| Chitin 1h | 5.6785 | 3.30225 | 0.960997 | 0.557107 |
| Epi 1h | 6.37027 | 3.28543 | 1.11377 | 0.585549 |
| SA 1h | 5.6105 | 3.25026 | 0.833433 | 0.482628 |
| Me-JA 1h | 10.3265 | 4.08456 | 1.67365 | 0.851072 |
| Control 6h | 11.1823 | 3.47441 | 1.61685 | 0.575822 |
| ABA 6h | 7.4938 | 3.54769 | 1.06797 | 0.514479 |
| ACC 6h | 7.62072 | 4.18237 | 0.992962 | 0.531126 |
| BABA 6h | 11.7045 | 3.88126 | 1.48997 | 0.565527 |
| Chitin 6h | 10.2771 | 3.76097 | 1.36759 | 0.582082 |
| Epi 6h | 6.72589 | 3.93228 | 0.903946 | 0.524539 |
| SA 6h | 7.58215 | 3.58417 | 1.14945 | 0.566358 |
| Me-JA 6h | 31.1645 | 25.3689 | 2.14663 | 3.11783 |
Source Transcript PGSC0003DMT400007381 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G33290.1 | +3 | 1e-146 | 434 | 216/302 (72%) | P-loop containing nucleoside triphosphate hydrolases superfamily protein | chr1:12074125-12075989 FORWARD LENGTH=379 |
