Probe CUST_11004_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11004_PI426222305 | JHI_St_60k_v1 | DMT400007382 | CTCTGTCTGATCTGGTATGTTTGGAACACCTAATGATTTCTTTAATGAGGAACTCCTCGT |
All Microarray Probes Designed to Gene DMG400002837
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11054_PI426222305 | JHI_St_60k_v1 | DMT400007385 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11283_PI426222305 | JHI_St_60k_v1 | DMT400007381 | AATAGCTCTTTTTCATTTTGGAGAACAAAATGCAGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11337_PI426222305 | JHI_St_60k_v1 | DMT400007384 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11004_PI426222305 | JHI_St_60k_v1 | DMT400007382 | CTCTGTCTGATCTGGTATGTTTGGAACACCTAATGATTTCTTTAATGAGGAACTCCTCGT |
Microarray Signals from CUST_11004_PI426222305
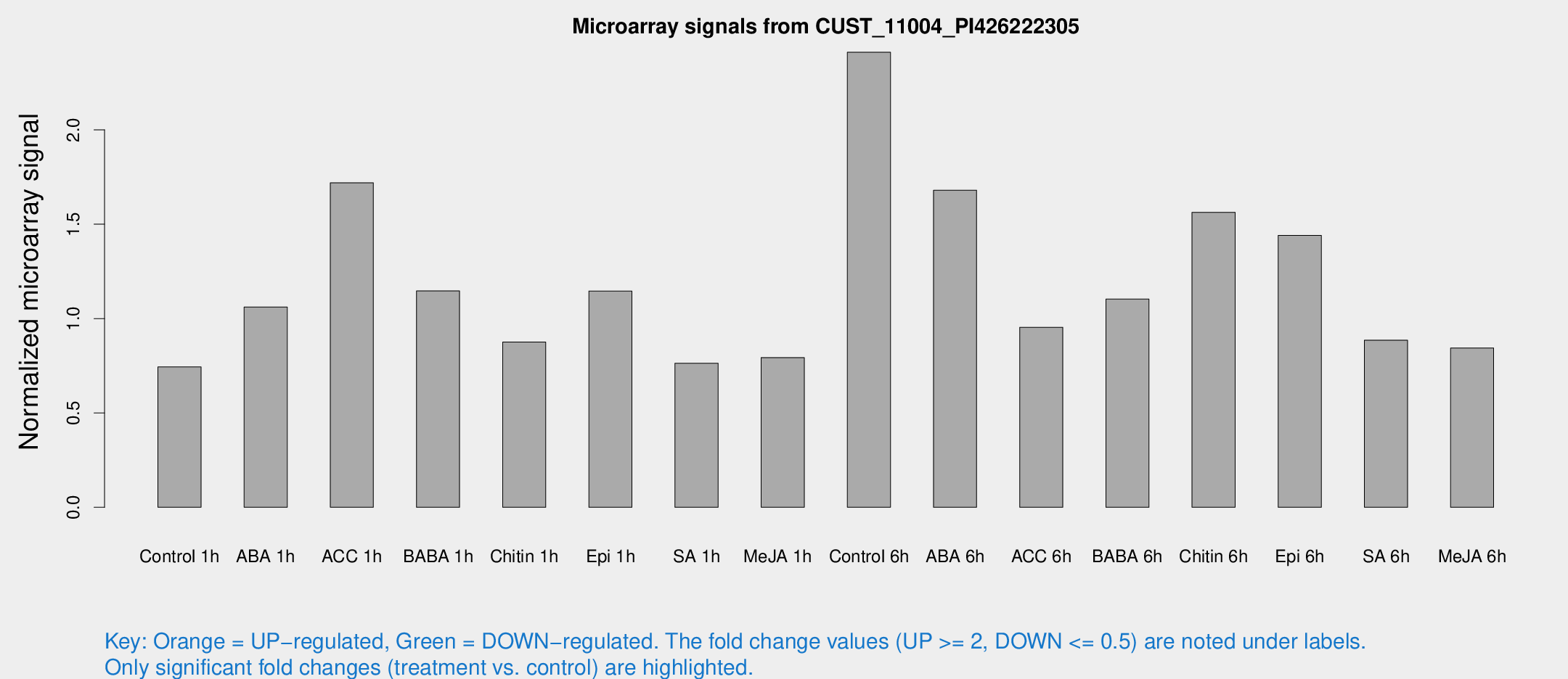
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.01793 | 2.8602 | 0.743937 | 0.366214 |
| ABA 1h | 7.65986 | 2.76379 | 1.06126 | 0.399101 |
| ACC 1h | 16.3621 | 5.30173 | 1.71903 | 0.633377 |
| BABA 1h | 9.94344 | 3.40688 | 1.14633 | 0.447333 |
| Chitin 1h | 6.42524 | 2.86156 | 0.875255 | 0.404577 |
| Epi 1h | 8.58523 | 2.87131 | 1.14559 | 0.452612 |
| SA 1h | 6.60673 | 2.89037 | 0.763144 | 0.372226 |
| Me-JA 1h | 4.99747 | 2.91075 | 0.79341 | 0.442377 |
| Control 6h | 19.8945 | 3.36498 | 2.411 | 0.402758 |
| ABA 6h | 14.5444 | 3.1635 | 1.67985 | 0.379907 |
| ACC 6h | 10.0805 | 3.86342 | 0.953885 | 0.39938 |
| BABA 6h | 10.8038 | 3.34799 | 1.1029 | 0.420196 |
| Chitin 6h | 13.7959 | 3.35215 | 1.56306 | 0.421092 |
| Epi 6h | 15.3742 | 6.27574 | 1.44098 | 0.725188 |
| SA 6h | 7.64601 | 3.14298 | 0.88525 | 0.427006 |
| Me-JA 6h | 6.85476 | 2.93711 | 0.844032 | 0.378058 |
Source Transcript PGSC0003DMT400007382 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G33290.1 | +3 | 1e-117 | 351 | 185/291 (64%) | P-loop containing nucleoside triphosphate hydrolases superfamily protein | chr1:12074125-12075989 FORWARD LENGTH=379 |
