Probe CUST_11054_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11054_PI426222305 | JHI_St_60k_v1 | DMT400007385 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
All Microarray Probes Designed to Gene DMG400002837
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_11054_PI426222305 | JHI_St_60k_v1 | DMT400007385 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11283_PI426222305 | JHI_St_60k_v1 | DMT400007381 | AATAGCTCTTTTTCATTTTGGAGAACAAAATGCAGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11337_PI426222305 | JHI_St_60k_v1 | DMT400007384 | AAATAAACGAGAGAGACTACTGGATTGTACACCGGACTGAAAGAAGTGTCGATGCTCTGC |
| CUST_11004_PI426222305 | JHI_St_60k_v1 | DMT400007382 | CTCTGTCTGATCTGGTATGTTTGGAACACCTAATGATTTCTTTAATGAGGAACTCCTCGT |
Microarray Signals from CUST_11054_PI426222305
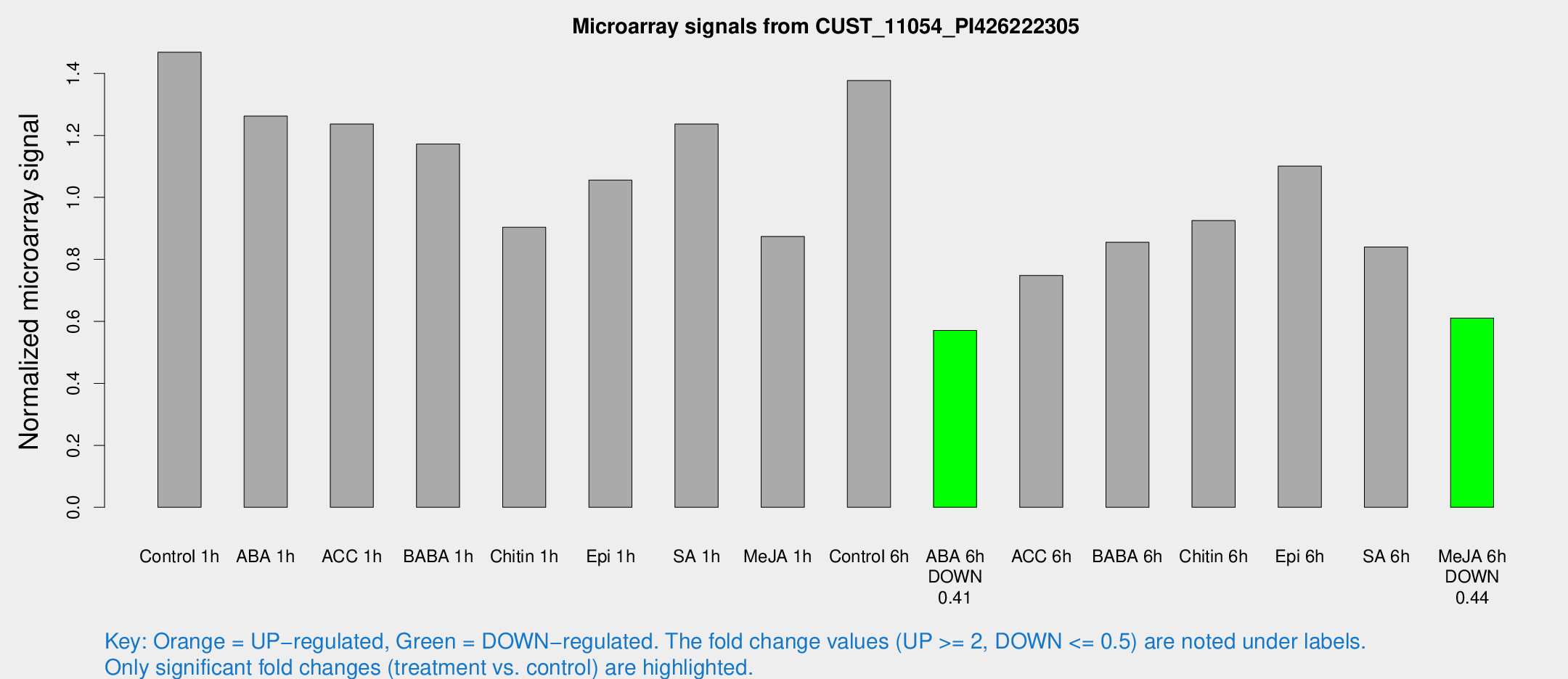
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 373.226 | 47.5693 | 1.46829 | 0.0957544 |
| ABA 1h | 287.153 | 44.0228 | 1.26218 | 0.23369 |
| ACC 1h | 327.328 | 52.762 | 1.2363 | 0.120402 |
| BABA 1h | 287.862 | 36.0362 | 1.17234 | 0.0697691 |
| Chitin 1h | 206.219 | 23.4918 | 0.903505 | 0.0782654 |
| Epi 1h | 233.268 | 31.0126 | 1.05585 | 0.141258 |
| SA 1h | 319.489 | 22.4856 | 1.23642 | 0.110033 |
| Me-JA 1h | 179.63 | 13.9297 | 0.873772 | 0.0535449 |
| Control 6h | 370.13 | 93.6197 | 1.37698 | 0.276433 |
| ABA 6h | 153.177 | 14.1831 | 0.57078 | 0.0346717 |
| ACC 6h | 225.785 | 50.3758 | 0.748207 | 0.0749321 |
| BABA 6h | 250.498 | 54.1644 | 0.854936 | 0.167864 |
| Chitin 6h | 246.997 | 14.8844 | 0.925181 | 0.0556904 |
| Epi 6h | 316.628 | 42.8534 | 1.10084 | 0.209615 |
| SA 6h | 215.853 | 42.3279 | 0.839848 | 0.101878 |
| Me-JA 6h | 157.724 | 27.8467 | 0.610376 | 0.0702994 |
Source Transcript PGSC0003DMT400007385 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G33290.1 | +1 | 1e-143 | 424 | 208/275 (76%) | P-loop containing nucleoside triphosphate hydrolases superfamily protein | chr1:12074125-12075989 FORWARD LENGTH=379 |
