Probe CUST_45902_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45902_PI426222305 | JHI_St_60k_v1 | DMT400048023 | GTTCCCCTCTACAGGGAATGCATTCATCCCCGAATTCCACAATTGGCAATAATTGGATTC |
All Microarray Probes Designed to Gene DMG400018659
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_45912_PI426222305 | JHI_St_60k_v1 | DMT400048020 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45941_PI426222305 | JHI_St_60k_v1 | DMT400048019 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45902_PI426222305 | JHI_St_60k_v1 | DMT400048023 | GTTCCCCTCTACAGGGAATGCATTCATCCCCGAATTCCACAATTGGCAATAATTGGATTC |
| CUST_45942_PI426222305 | JHI_St_60k_v1 | DMT400048021 | GGATCAATTACTTGCTCATATGGTAATTGGTATAGGACACGTGTTATTCAACTGCAAGGC |
| CUST_45900_PI426222305 | JHI_St_60k_v1 | DMT400048022 | CTCAGATGATTCTTCAGTTCCCCTCTACAGGTTAGTAATAATTTCTGCACCAAATTAATT |
Microarray Signals from CUST_45902_PI426222305
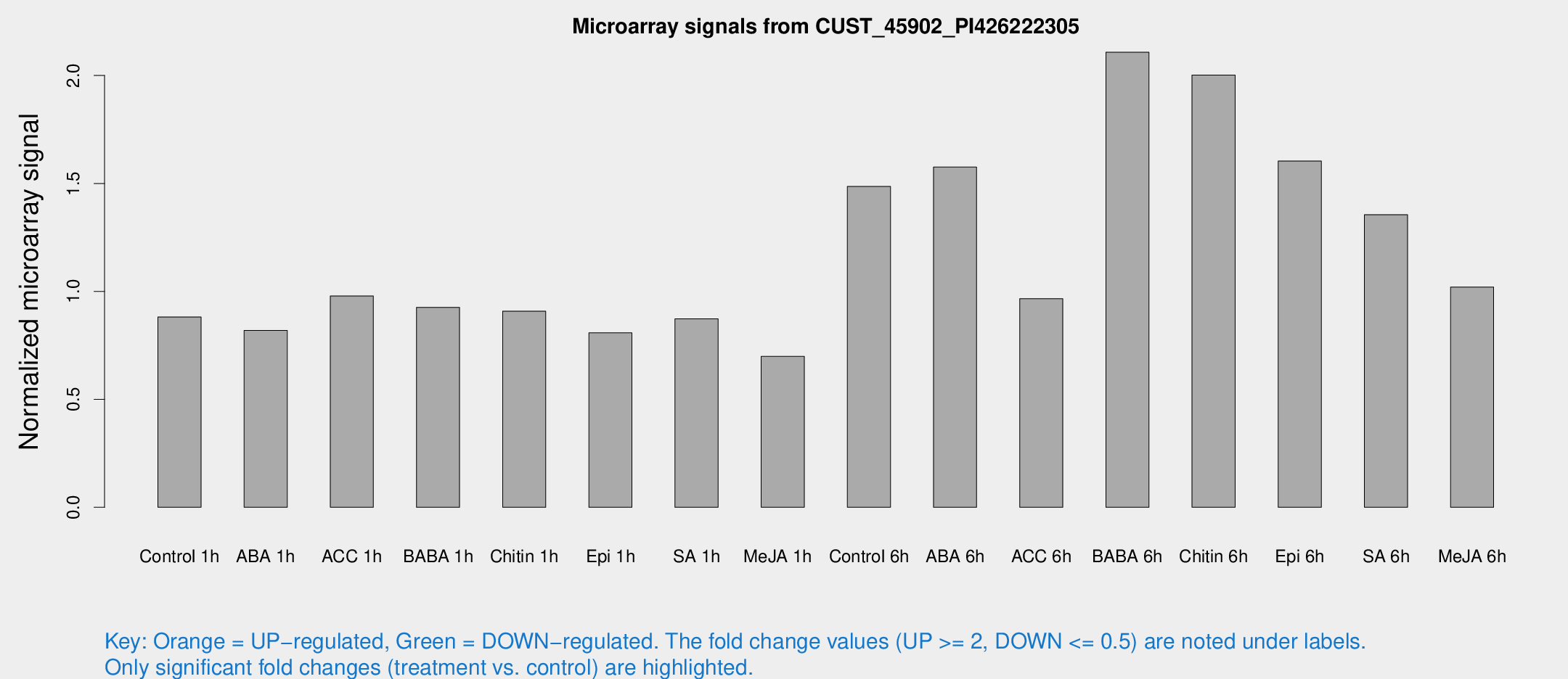
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 223.043 | 31.7267 | 0.881654 | 0.0649961 |
| ABA 1h | 182.107 | 20.9736 | 0.819182 | 0.0493807 |
| ACC 1h | 253.589 | 33.2279 | 0.978651 | 0.0719479 |
| BABA 1h | 222.839 | 16.9417 | 0.92605 | 0.0553069 |
| Chitin 1h | 205.927 | 24.8581 | 0.908369 | 0.0557487 |
| Epi 1h | 175.232 | 17.2397 | 0.808142 | 0.0592509 |
| SA 1h | 222.597 | 13.2517 | 0.873067 | 0.0677817 |
| Me-JA 1h | 142.391 | 10.9698 | 0.698885 | 0.114505 |
| Control 6h | 433.148 | 140.349 | 1.48603 | 0.554589 |
| ABA 6h | 418.319 | 34.1502 | 1.57566 | 0.204887 |
| ACC 6h | 288.659 | 62.8523 | 0.966323 | 0.100931 |
| BABA 6h | 587.592 | 35.5454 | 2.10771 | 0.176791 |
| Chitin 6h | 529.859 | 30.8709 | 2.00163 | 0.116376 |
| Epi 6h | 454.279 | 51.8771 | 1.6039 | 0.200322 |
| SA 6h | 336.135 | 34.0482 | 1.35533 | 0.0795991 |
| Me-JA 6h | 270.794 | 66.4658 | 1.02028 | 0.203783 |
Source Transcript PGSC0003DMT400048023 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19250.1 | +1 | 5e-111 | 346 | 206/499 (41%) | flavin-dependent monooxygenase 1 | chr1:6650656-6653053 REVERSE LENGTH=530 |
