Probe CUST_17945_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17945_PI426222305 | JHI_St_60k_v1 | DMT400018219 | CTGATCATAATTTAGCACCAAGAATGGTGCCTTTCTTTCTTGAAACCATCAAGAAATATG |
All Microarray Probes Designed to Gene DMG400007074
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17863_PI426222305 | JHI_St_60k_v1 | DMT400018226 | GACAAATGGGCAAAACATAGAAAAATCATCAATCCCGCGTTCCATCTAGAGAAGTTAAAG |
| CUST_18057_PI426222305 | JHI_St_60k_v1 | DMT400018220 | AGTGCTTGGGAGTCGGAAACCAGATTTTGATGGATTAAATCATCTAAAAGTTGCAAAAAA |
| CUST_17984_PI426222305 | JHI_St_60k_v1 | DMT400018224 | GTCTTGCAAGTGTTTGGGAGTCGGAAACCAGATTTTGATGGCTTAAATCATCTAAAAGTT |
| CUST_18102_PI426222305 | JHI_St_60k_v1 | DMT400018222 | CTGATCATAATTTAGCACCAAGAATGGTGCCTTTCTTTCTTGAAACCATCAAGAAATATG |
| CUST_18070_PI426222305 | JHI_St_60k_v1 | DMT400018225 | CGTGCTAGCACGAGCCTAATATATGTTGCTCTCTTAATATCAAATTATGTGATATAGTTG |
| CUST_17945_PI426222305 | JHI_St_60k_v1 | DMT400018219 | CTGATCATAATTTAGCACCAAGAATGGTGCCTTTCTTTCTTGAAACCATCAAGAAATATG |
| CUST_17891_PI426222305 | JHI_St_60k_v1 | DMT400018223 | AAGTGTTAGTGTTATTGACATTTTCCCTGGTTCACCATTTTCTCAAACAGCATATGCTTC |
Microarray Signals from CUST_17945_PI426222305
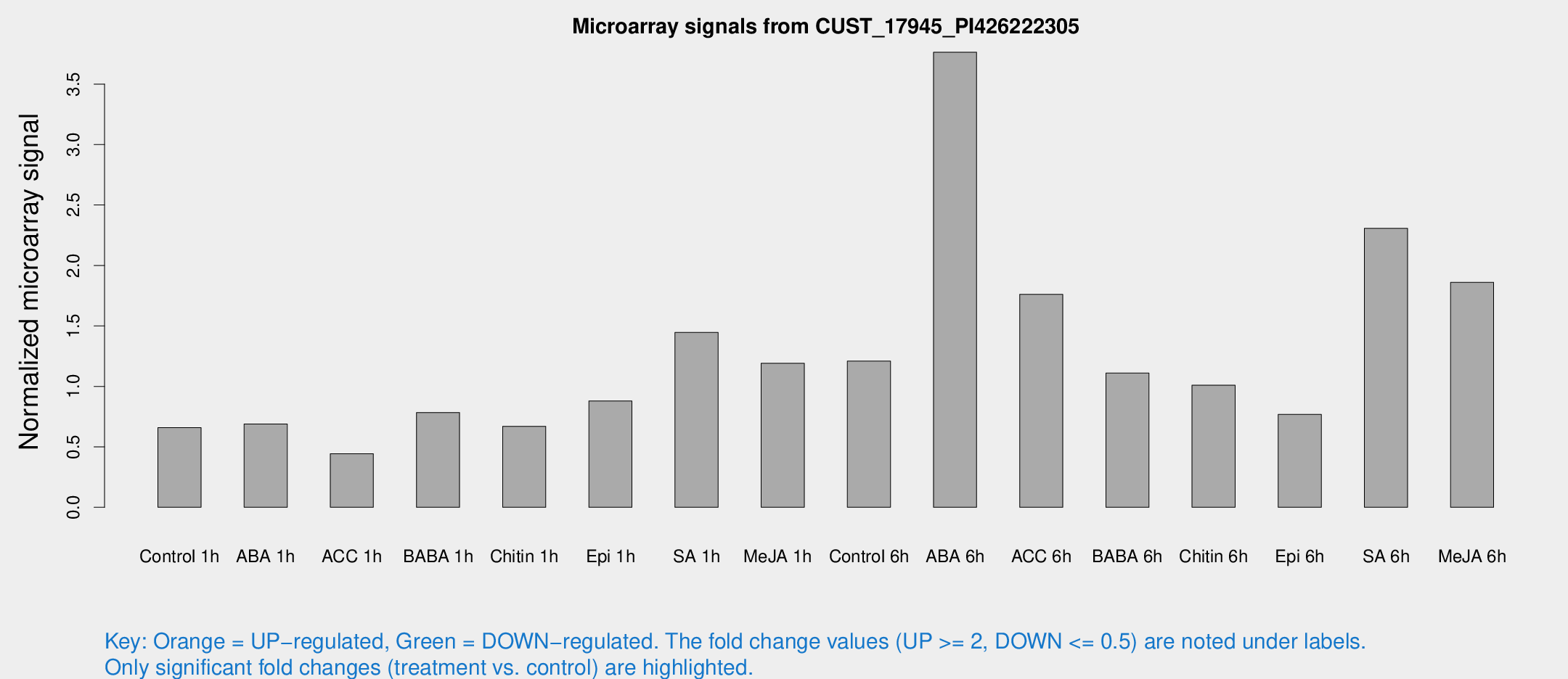
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 77.8305 | 7.29147 | 0.659204 | 0.0758435 |
| ABA 1h | 73.5584 | 13.3344 | 0.688649 | 0.144173 |
| ACC 1h | 64.513 | 26.1699 | 0.442363 | 0.195689 |
| BABA 1h | 95.7975 | 24.143 | 0.783395 | 0.156529 |
| Chitin 1h | 71.9561 | 10.3397 | 0.6697 | 0.049643 |
| Epi 1h | 89.8264 | 8.15371 | 0.879419 | 0.0858518 |
| SA 1h | 186.976 | 44.4705 | 1.44656 | 0.353197 |
| Me-JA 1h | 125.893 | 39.6353 | 1.19128 | 0.285818 |
| Control 6h | 155.658 | 43.9038 | 1.20899 | 0.278829 |
| ABA 6h | 555.658 | 191.332 | 3.76331 | 1.70042 |
| ACC 6h | 237.879 | 18.39 | 1.7608 | 0.338636 |
| BABA 6h | 155.984 | 36.8016 | 1.10955 | 0.277524 |
| Chitin 6h | 127.128 | 12.4016 | 1.00953 | 0.111279 |
| Epi 6h | 107.655 | 26.1574 | 0.768553 | 0.22768 |
| SA 6h | 350.675 | 134.911 | 2.3067 | 2.24035 |
| Me-JA 6h | 219.666 | 25.1415 | 1.86041 | 0.166646 |
Source Transcript PGSC0003DMT400018219 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G14690.1 | +2 | 0.0 | 617 | 295/518 (57%) | cytochrome P450, family 72, subfamily A, polypeptide 15 | chr3:4937410-4939310 FORWARD LENGTH=512 |
