Probe CUST_14453_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14453_PI426222305 | JHI_St_60k_v1 | DMT400066425 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
All Microarray Probes Designed to Gene DMG401025828
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_14453_PI426222305 | JHI_St_60k_v1 | DMT400066425 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14484_PI426222305 | JHI_St_60k_v1 | DMT400066422 | CCCCATTTAAAAGCAGCAGCAATTATGTTCAAAAGATAGGTCTTCTATTCTGAGTTTTCT |
| CUST_14615_PI426222305 | JHI_St_60k_v1 | DMT400066420 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14555_PI426222305 | JHI_St_60k_v1 | DMT400066421 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
| CUST_14729_PI426222305 | JHI_St_60k_v1 | DMT400066419 | TTGGGAGCATGGTTCTTAATTTTGAGTTGCACTTTAGCCAATGTATTCTAGATAAGGTAT |
Microarray Signals from CUST_14453_PI426222305
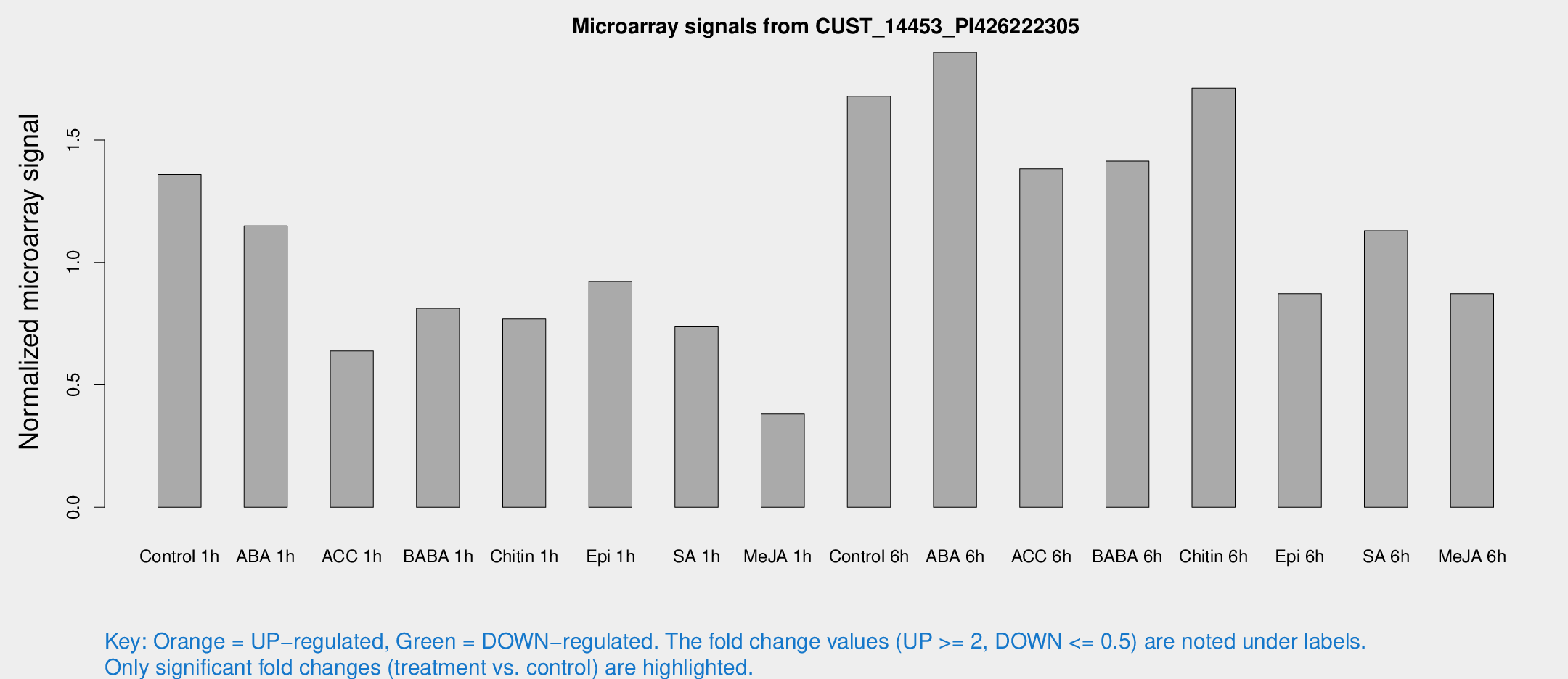
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 52.7798 | 8.45863 | 1.35914 | 0.21888 |
| ABA 1h | 41.4334 | 10.8945 | 1.1498 | 0.296815 |
| ACC 1h | 30.7947 | 13.4908 | 0.638537 | 0.302114 |
| BABA 1h | 30.8166 | 5.49016 | 0.812748 | 0.123825 |
| Chitin 1h | 26.9181 | 4.1235 | 0.769117 | 0.116269 |
| Epi 1h | 30.547 | 3.97936 | 0.922255 | 0.124336 |
| SA 1h | 29.4902 | 4.98735 | 0.736948 | 0.141079 |
| Me-JA 1h | 11.8548 | 3.61118 | 0.380884 | 0.11774 |
| Control 6h | 68.0139 | 15.4253 | 1.6786 | 0.30029 |
| ABA 6h | 85.6523 | 25.95 | 1.85855 | 0.721573 |
| ACC 6h | 60.5313 | 5.57674 | 1.38268 | 0.123878 |
| BABA 6h | 64.8204 | 15.9356 | 1.41411 | 0.382869 |
| Chitin 6h | 72.3926 | 15.9843 | 1.71256 | 0.335971 |
| Epi 6h | 43.0987 | 17.0656 | 0.872636 | 0.371779 |
| SA 6h | 51.8007 | 23.3549 | 1.12957 | 0.814493 |
| Me-JA 6h | 36.0792 | 9.84979 | 0.872974 | 0.209337 |
Source Transcript PGSC0003DMT400066425 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65940.1 | +3 | 3e-75 | 242 | 117/155 (75%) | beta-hydroxyisobutyryl-CoA hydrolase 1 | chr5:26376830-26379161 REVERSE LENGTH=378 |
