Probe CUST_9915_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9915_PI426222305 | JHI_St_60k_v1 | DMT400065328 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
All Microarray Probes Designed to Gene DMG400025395
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9966_PI426222305 | JHI_St_60k_v1 | DMT400065325 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9915_PI426222305 | JHI_St_60k_v1 | DMT400065328 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9949_PI426222305 | JHI_St_60k_v1 | DMT400065329 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9960_PI426222305 | JHI_St_60k_v1 | DMT400065327 | GCTGGTCGATTTTGGACTATTATTGTTGCTTCCATTATTTATGAAATGGTACGTCTCTAT |
| CUST_9977_PI426222305 | JHI_St_60k_v1 | DMT400065326 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
Microarray Signals from CUST_9915_PI426222305
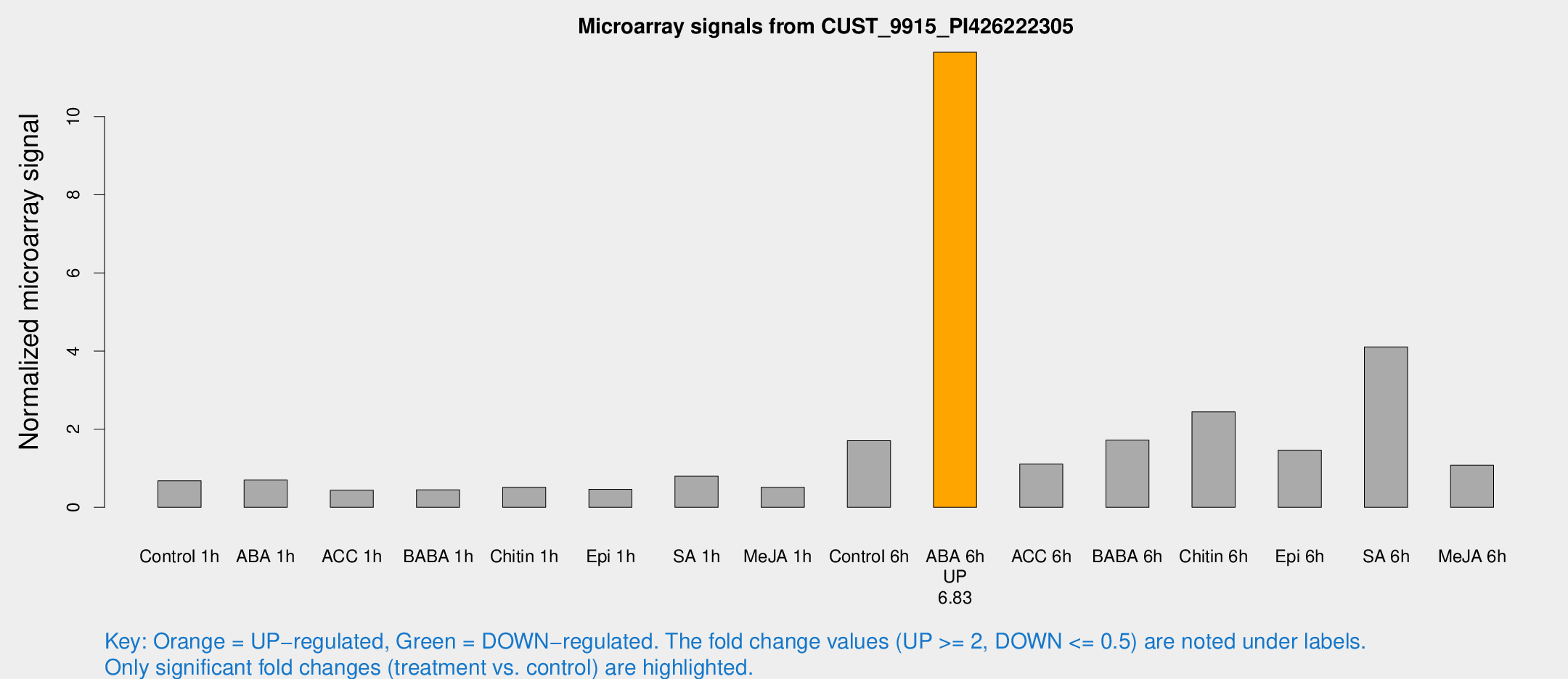
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 97.0849 | 27.5025 | 0.677553 | 0.148274 |
| ABA 1h | 83.8737 | 11.8667 | 0.696869 | 0.156967 |
| ACC 1h | 60.0956 | 4.9334 | 0.436391 | 0.0357965 |
| BABA 1h | 62.3111 | 16.3211 | 0.449095 | 0.0889385 |
| Chitin 1h | 62.9419 | 9.71268 | 0.512411 | 0.0669058 |
| Epi 1h | 57.7173 | 16.7635 | 0.461773 | 0.123536 |
| SA 1h | 115.718 | 28.7532 | 0.798791 | 0.241444 |
| Me-JA 1h | 55.5914 | 4.57764 | 0.510469 | 0.0624317 |
| Control 6h | 251.8 | 68.3687 | 1.70497 | 0.438936 |
| ABA 6h | 1727.76 | 347.52 | 11.6473 | 2.02879 |
| ACC 6h | 176.336 | 33.6589 | 1.10627 | 0.154915 |
| BABA 6h | 280.654 | 81.5629 | 1.71894 | 0.602348 |
| Chitin 6h | 395.903 | 150.304 | 2.44278 | 0.944977 |
| Epi 6h | 223.846 | 30.0444 | 1.46416 | 0.317153 |
| SA 6h | 551.841 | 74.8631 | 4.10439 | 0.924061 |
| Me-JA 6h | 144.309 | 14.5403 | 1.07648 | 0.0672004 |
Source Transcript PGSC0003DMT400065328 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G68570.1 | +3 | 4e-161 | 485 | 252/469 (54%) | Major facilitator superfamily protein | chr1:25746811-25750110 FORWARD LENGTH=596 |
