Probe CUST_9966_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9966_PI426222305 | JHI_St_60k_v1 | DMT400065325 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
All Microarray Probes Designed to Gene DMG400025395
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9966_PI426222305 | JHI_St_60k_v1 | DMT400065325 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9915_PI426222305 | JHI_St_60k_v1 | DMT400065328 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9949_PI426222305 | JHI_St_60k_v1 | DMT400065329 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9960_PI426222305 | JHI_St_60k_v1 | DMT400065327 | GCTGGTCGATTTTGGACTATTATTGTTGCTTCCATTATTTATGAAATGGTACGTCTCTAT |
| CUST_9977_PI426222305 | JHI_St_60k_v1 | DMT400065326 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
Microarray Signals from CUST_9966_PI426222305
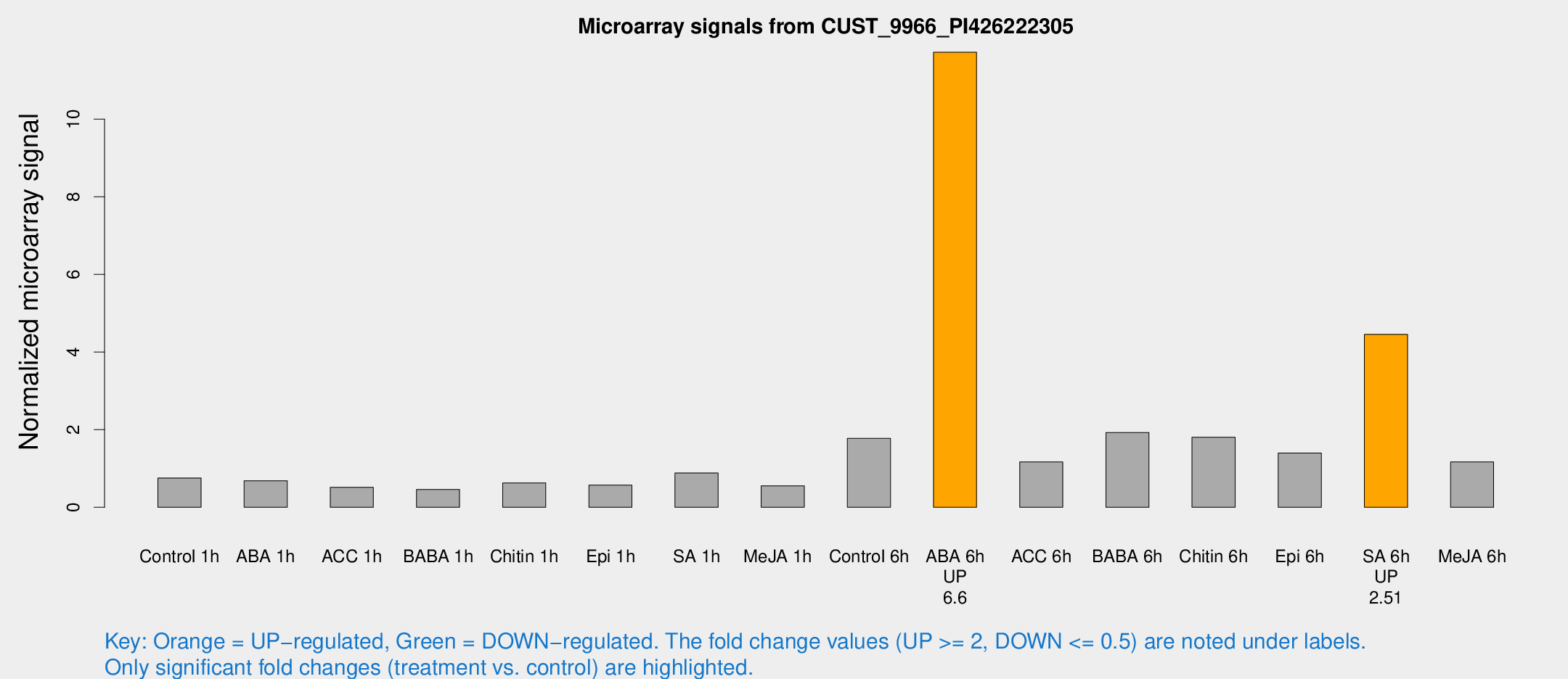
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 104.656 | 21.5845 | 0.754854 | 0.114862 |
| ABA 1h | 82.7774 | 12.5513 | 0.684538 | 0.165304 |
| ACC 1h | 73.7861 | 15.4631 | 0.515897 | 0.0824083 |
| BABA 1h | 62.2403 | 12.7044 | 0.460989 | 0.0600024 |
| Chitin 1h | 76.7306 | 9.7378 | 0.629557 | 0.136987 |
| Epi 1h | 67.3465 | 9.80937 | 0.572137 | 0.0661695 |
| SA 1h | 131.793 | 39.6957 | 0.882939 | 0.323023 |
| Me-JA 1h | 60.7104 | 5.61334 | 0.553451 | 0.0468354 |
| Control 6h | 256.807 | 63.6895 | 1.77536 | 0.371676 |
| ABA 6h | 1722.42 | 319.471 | 11.7242 | 1.63586 |
| ACC 6h | 183.409 | 27.9951 | 1.16951 | 0.109227 |
| BABA 6h | 313.98 | 88.3347 | 1.92804 | 0.672955 |
| Chitin 6h | 259.104 | 27.9755 | 1.8059 | 0.15469 |
| Epi 6h | 214.694 | 29.2099 | 1.39933 | 0.306925 |
| SA 6h | 589.365 | 34.3611 | 4.4559 | 0.752705 |
| Me-JA 6h | 156.987 | 16.9703 | 1.16934 | 0.0731032 |
Source Transcript PGSC0003DMT400065325 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G68570.1 | +2 | 1e-98 | 322 | 156/283 (55%) | Major facilitator superfamily protein | chr1:25746811-25750110 FORWARD LENGTH=596 |
