Probe CUST_9960_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9960_PI426222305 | JHI_St_60k_v1 | DMT400065327 | GCTGGTCGATTTTGGACTATTATTGTTGCTTCCATTATTTATGAAATGGTACGTCTCTAT |
All Microarray Probes Designed to Gene DMG400025395
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_9966_PI426222305 | JHI_St_60k_v1 | DMT400065325 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9915_PI426222305 | JHI_St_60k_v1 | DMT400065328 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9949_PI426222305 | JHI_St_60k_v1 | DMT400065329 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
| CUST_9960_PI426222305 | JHI_St_60k_v1 | DMT400065327 | GCTGGTCGATTTTGGACTATTATTGTTGCTTCCATTATTTATGAAATGGTACGTCTCTAT |
| CUST_9977_PI426222305 | JHI_St_60k_v1 | DMT400065326 | CTTTGTTAGACAATTCTAAAGGTGATGGACAAACAGATAGAGCAAGAAATGGAATGAACT |
Microarray Signals from CUST_9960_PI426222305
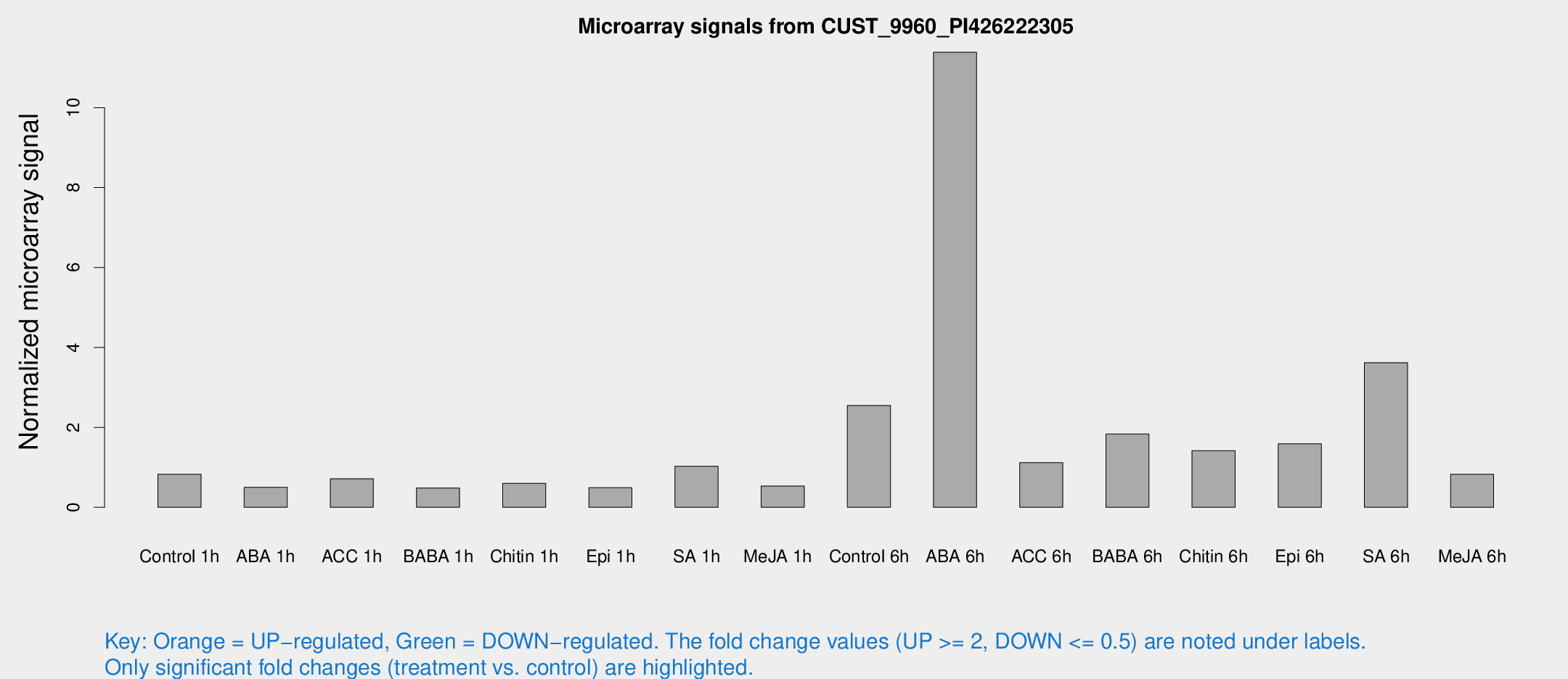
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.9741 | 3.53063 | 0.825899 | 0.297471 |
| ABA 1h | 5.70515 | 3.07232 | 0.498468 | 0.270846 |
| ACC 1h | 9.60695 | 3.75642 | 0.714314 | 0.296364 |
| BABA 1h | 5.98283 | 3.47288 | 0.48072 | 0.278429 |
| Chitin 1h | 7.19917 | 3.23603 | 0.601992 | 0.293778 |
| Epi 1h | 5.44459 | 3.16567 | 0.490383 | 0.282488 |
| SA 1h | 14.4457 | 3.30601 | 1.02887 | 0.34726 |
| Me-JA 1h | 5.63652 | 3.28866 | 0.532385 | 0.308448 |
| Control 6h | 38.3227 | 12.8457 | 2.54699 | 0.923881 |
| ABA 6h | 175.465 | 51.2709 | 11.3873 | 3.94666 |
| ACC 6h | 16.782 | 4.13727 | 1.11563 | 0.276076 |
| BABA 6h | 28.6898 | 8.3131 | 1.83344 | 0.50791 |
| Chitin 6h | 19.732 | 3.86496 | 1.41599 | 0.29243 |
| Epi 6h | 24.7083 | 5.74724 | 1.58995 | 0.596699 |
| SA 6h | 58.7572 | 21.9278 | 3.62039 | 3.18858 |
| Me-JA 6h | 11.6806 | 3.36452 | 0.826455 | 0.301149 |
Source Transcript PGSC0003DMT400065327 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G68570.1 | +1 | 3e-30 | 117 | 52/83 (63%) | Major facilitator superfamily protein | chr1:25746811-25750110 FORWARD LENGTH=596 |
