Probe CUST_8751_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8751_PI426222305 | JHI_St_60k_v1 | DMT400046382 | GAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTTTTT |
All Microarray Probes Designed to Gene DMG400018004
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8784_PI426222305 | JHI_St_60k_v1 | DMT400046381 | GAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTTTTT |
| CUST_8700_PI426222305 | JHI_St_60k_v1 | DMT400046380 | CTGAGTCAAACACTTACAGCCCATTTATATGGGGTTTATGATGGACATGGAGGCTCTCAG |
| CUST_8751_PI426222305 | JHI_St_60k_v1 | DMT400046382 | GAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTTTTT |
| CUST_8709_PI426222305 | JHI_St_60k_v1 | DMT400046379 | GAAGAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTT |
| CUST_8774_PI426222305 | JHI_St_60k_v1 | DMT400046378 | GAAGAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTT |
Microarray Signals from CUST_8751_PI426222305
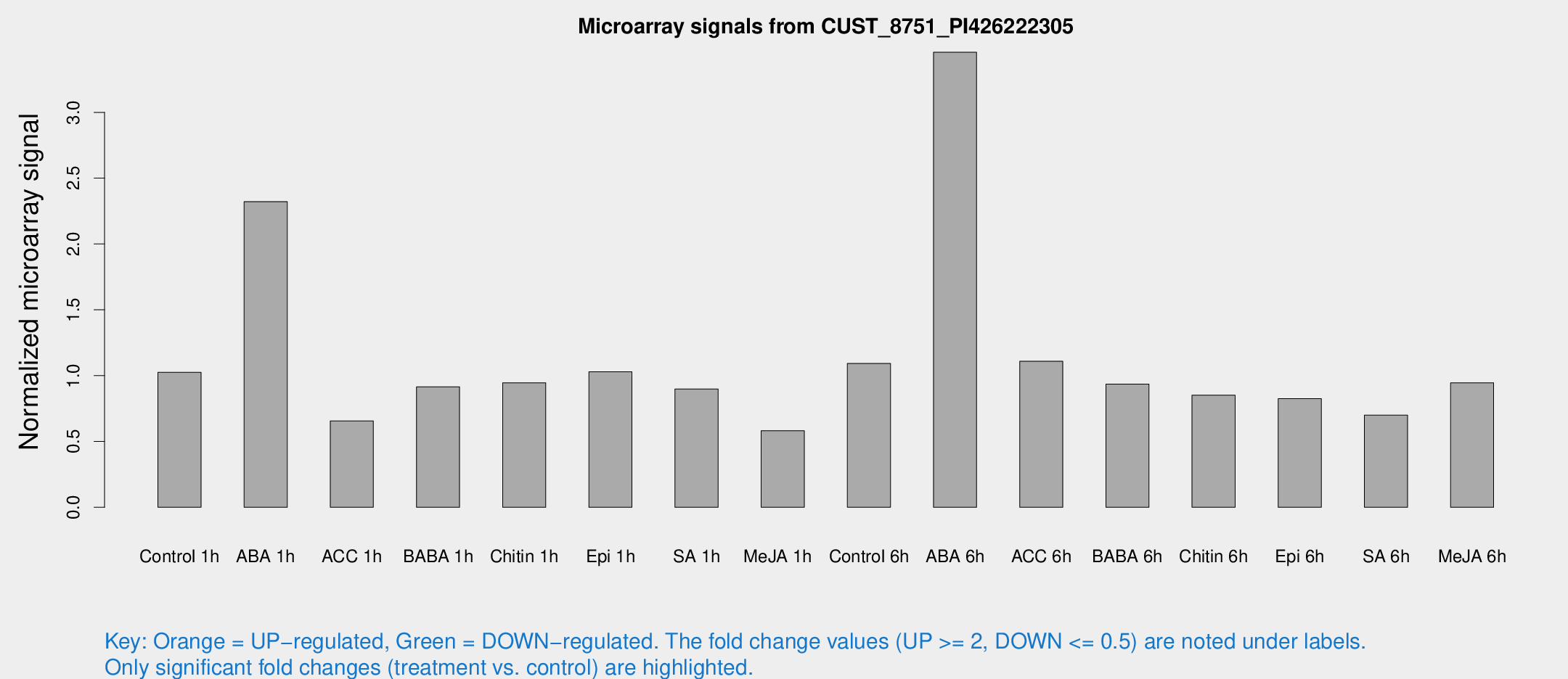
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 76.28 | 10.6175 | 1.02557 | 0.116422 |
| ABA 1h | 155.198 | 30.3638 | 2.32213 | 0.433724 |
| ACC 1h | 56.1778 | 17.7655 | 0.656548 | 0.226455 |
| BABA 1h | 66.6618 | 13.3674 | 0.915354 | 0.150858 |
| Chitin 1h | 62.4772 | 5.68582 | 0.945832 | 0.0733233 |
| Epi 1h | 65.3213 | 5.36417 | 1.02912 | 0.0815267 |
| SA 1h | 68.3081 | 8.13155 | 0.899055 | 0.138707 |
| Me-JA 1h | 39.0568 | 13.7895 | 0.582147 | 0.158017 |
| Control 6h | 83.8023 | 16.8685 | 1.09275 | 0.152927 |
| ABA 6h | 299.792 | 85.8469 | 3.45666 | 1.17388 |
| ACC 6h | 96.0402 | 17.2956 | 1.10949 | 0.118852 |
| BABA 6h | 80.7374 | 17.1636 | 0.936428 | 0.203596 |
| Chitin 6h | 75.0675 | 22.4035 | 0.852262 | 0.369333 |
| Epi 6h | 70.8709 | 15.4404 | 0.826129 | 0.167983 |
| SA 6h | 59.9553 | 23.8021 | 0.699707 | 0.36761 |
| Me-JA 6h | 82.6795 | 32.1011 | 0.945705 | 0.390734 |
Source Transcript PGSC0003DMT400046382 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G57050.1 | +1 | 1e-118 | 365 | 200/345 (58%) | Protein phosphatase 2C family protein | chr5:23087720-23089303 FORWARD LENGTH=423 |
