Probe CUST_8700_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8700_PI426222305 | JHI_St_60k_v1 | DMT400046380 | CTGAGTCAAACACTTACAGCCCATTTATATGGGGTTTATGATGGACATGGAGGCTCTCAG |
All Microarray Probes Designed to Gene DMG400018004
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8784_PI426222305 | JHI_St_60k_v1 | DMT400046381 | GAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTTTTT |
| CUST_8700_PI426222305 | JHI_St_60k_v1 | DMT400046380 | CTGAGTCAAACACTTACAGCCCATTTATATGGGGTTTATGATGGACATGGAGGCTCTCAG |
| CUST_8751_PI426222305 | JHI_St_60k_v1 | DMT400046382 | GAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTTTTT |
| CUST_8709_PI426222305 | JHI_St_60k_v1 | DMT400046379 | GAAGAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTT |
| CUST_8774_PI426222305 | JHI_St_60k_v1 | DMT400046378 | GAAGAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTT |
Microarray Signals from CUST_8700_PI426222305
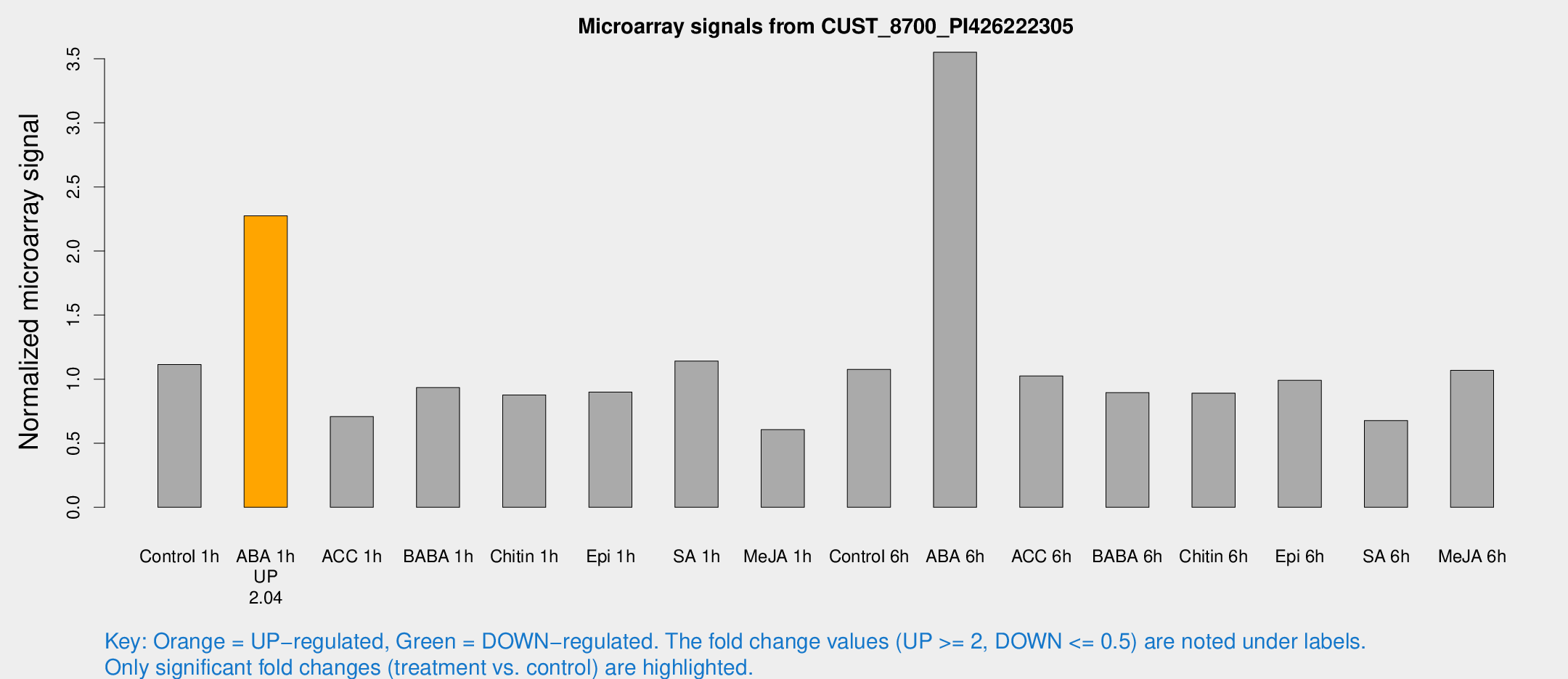
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 530.37 | 77.828 | 1.11387 | 0.100999 |
| ABA 1h | 954.225 | 124.071 | 2.27488 | 0.20807 |
| ACC 1h | 376.77 | 105.101 | 0.708468 | 0.205014 |
| BABA 1h | 454.284 | 116.283 | 0.934409 | 0.187706 |
| Chitin 1h | 389.433 | 99.8237 | 0.876788 | 0.160044 |
| Epi 1h | 365.572 | 33.9652 | 0.899328 | 0.076856 |
| SA 1h | 571.454 | 127.079 | 1.14137 | 0.177346 |
| Me-JA 1h | 236.341 | 38.6102 | 0.606576 | 0.0458039 |
| Control 6h | 558.229 | 159.077 | 1.0763 | 0.292111 |
| ABA 6h | 1914.97 | 488.559 | 3.551 | 1.00149 |
| ACC 6h | 557.702 | 80.5001 | 1.02446 | 0.0595254 |
| BABA 6h | 476.683 | 69.9389 | 0.895318 | 0.112743 |
| Chitin 6h | 446.943 | 50.4274 | 0.890203 | 0.100682 |
| Epi 6h | 545.016 | 123.923 | 0.990702 | 0.211955 |
| SA 6h | 400.42 | 152.069 | 0.676539 | 0.355319 |
| Me-JA 6h | 543.251 | 149.2 | 1.06867 | 0.262257 |
Source Transcript PGSC0003DMT400046380 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G57050.1 | +2 | 3e-64 | 224 | 125/238 (53%) | Protein phosphatase 2C family protein | chr5:23087720-23089303 FORWARD LENGTH=423 |
