Probe CUST_8709_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8709_PI426222305 | JHI_St_60k_v1 | DMT400046379 | GAAGAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTT |
All Microarray Probes Designed to Gene DMG400018004
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_8784_PI426222305 | JHI_St_60k_v1 | DMT400046381 | GAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTTTTT |
| CUST_8700_PI426222305 | JHI_St_60k_v1 | DMT400046380 | CTGAGTCAAACACTTACAGCCCATTTATATGGGGTTTATGATGGACATGGAGGCTCTCAG |
| CUST_8751_PI426222305 | JHI_St_60k_v1 | DMT400046382 | GAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTTTTT |
| CUST_8709_PI426222305 | JHI_St_60k_v1 | DMT400046379 | GAAGAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTT |
| CUST_8774_PI426222305 | JHI_St_60k_v1 | DMT400046378 | GAAGAAGTTGGGTTGCTTTGAATTTGGAAAAAAGTTTCATGTTTGGTCTGCTTTGAGCTT |
Microarray Signals from CUST_8709_PI426222305
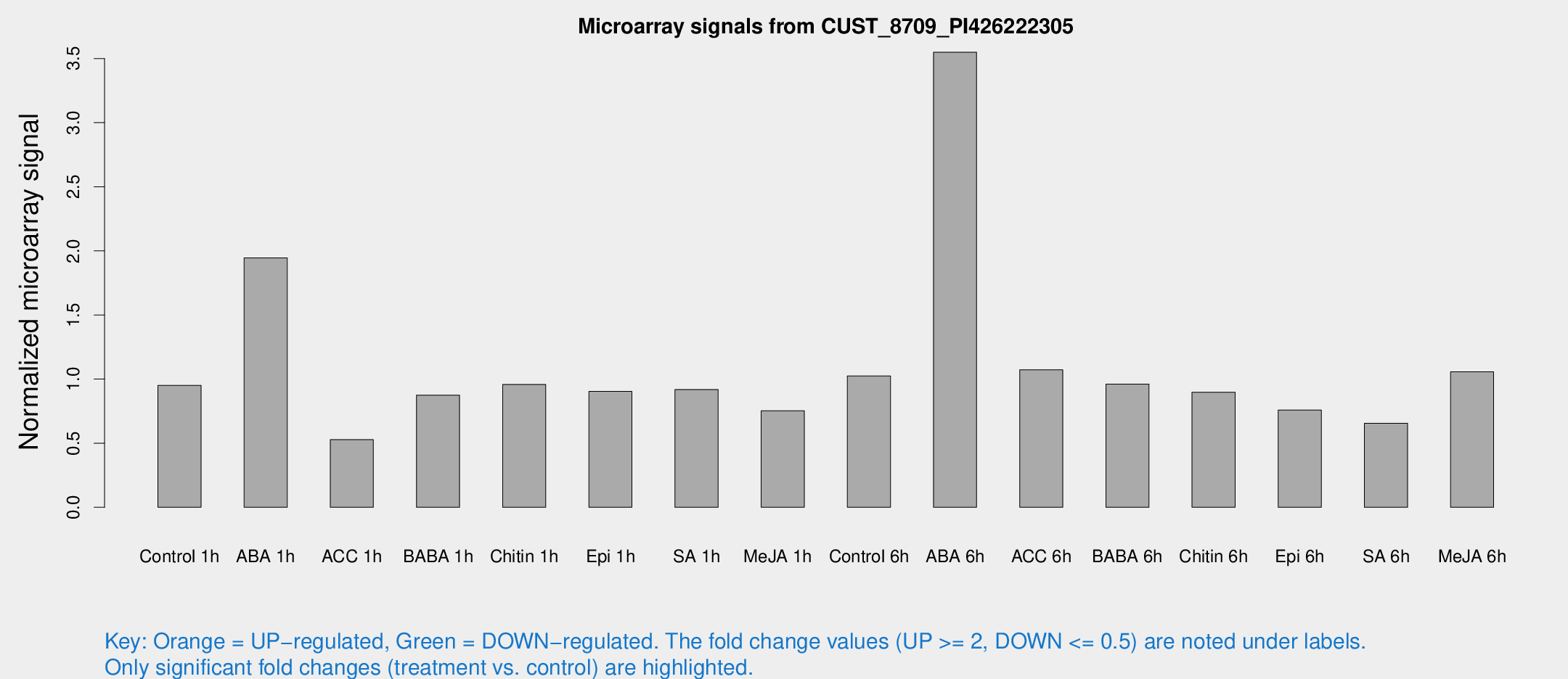
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 63.0115 | 8.91794 | 0.951581 | 0.125833 |
| ABA 1h | 117.681 | 26.7821 | 1.94477 | 0.42165 |
| ACC 1h | 41.9515 | 15.5254 | 0.527393 | 0.221744 |
| BABA 1h | 61.8825 | 18.6261 | 0.87484 | 0.250069 |
| Chitin 1h | 56.9171 | 7.10399 | 0.958207 | 0.0779361 |
| Epi 1h | 51.1417 | 4.32753 | 0.903913 | 0.086614 |
| SA 1h | 62.7511 | 9.32246 | 0.918716 | 0.0880229 |
| Me-JA 1h | 41.648 | 7.80258 | 0.753367 | 0.176686 |
| Control 6h | 70.1536 | 14.5241 | 1.02388 | 0.153066 |
| ABA 6h | 264.665 | 65.1773 | 3.54934 | 0.907043 |
| ACC 6h | 81.7269 | 12.9247 | 1.07198 | 0.0810656 |
| BABA 6h | 77.3518 | 22.6494 | 0.961308 | 0.299515 |
| Chitin 6h | 67.6317 | 19.7663 | 0.897758 | 0.30126 |
| Epi 6h | 59.1838 | 14.6616 | 0.759201 | 0.234484 |
| SA 6h | 52.8708 | 25.3288 | 0.655234 | 0.364899 |
| Me-JA 6h | 75.6601 | 24.0578 | 1.05667 | 0.260175 |
Source Transcript PGSC0003DMT400046379 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G57050.1 | +2 | 1e-64 | 224 | 125/238 (53%) | Protein phosphatase 2C family protein | chr5:23087720-23089303 FORWARD LENGTH=423 |
