Probe CUST_5782_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5782_PI426222305 | JHI_St_60k_v1 | DMT400022917 | ATTGAATACGTGACGATTCCCCCAAAAATAGTGTGTTTTTGGAGGATCCAACACGGGGCT |
All Microarray Probes Designed to Gene DMG400008882
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5701_PI426222305 | JHI_St_60k_v1 | DMT400022916 | TGCCATTTGTACTTCTAACATTTGAGTATGAGTTATTCCCTTCCATATATGGTCTCTCAT |
| CUST_5782_PI426222305 | JHI_St_60k_v1 | DMT400022917 | ATTGAATACGTGACGATTCCCCCAAAAATAGTGTGTTTTTGGAGGATCCAACACGGGGCT |
| CUST_5629_PI426222305 | JHI_St_60k_v1 | DMT400022914 | TGCCATTTGTACTTCTAACATTTGAGTATGAGTTATTCCCTTCCATATATGGTCTCTCAT |
Microarray Signals from CUST_5782_PI426222305
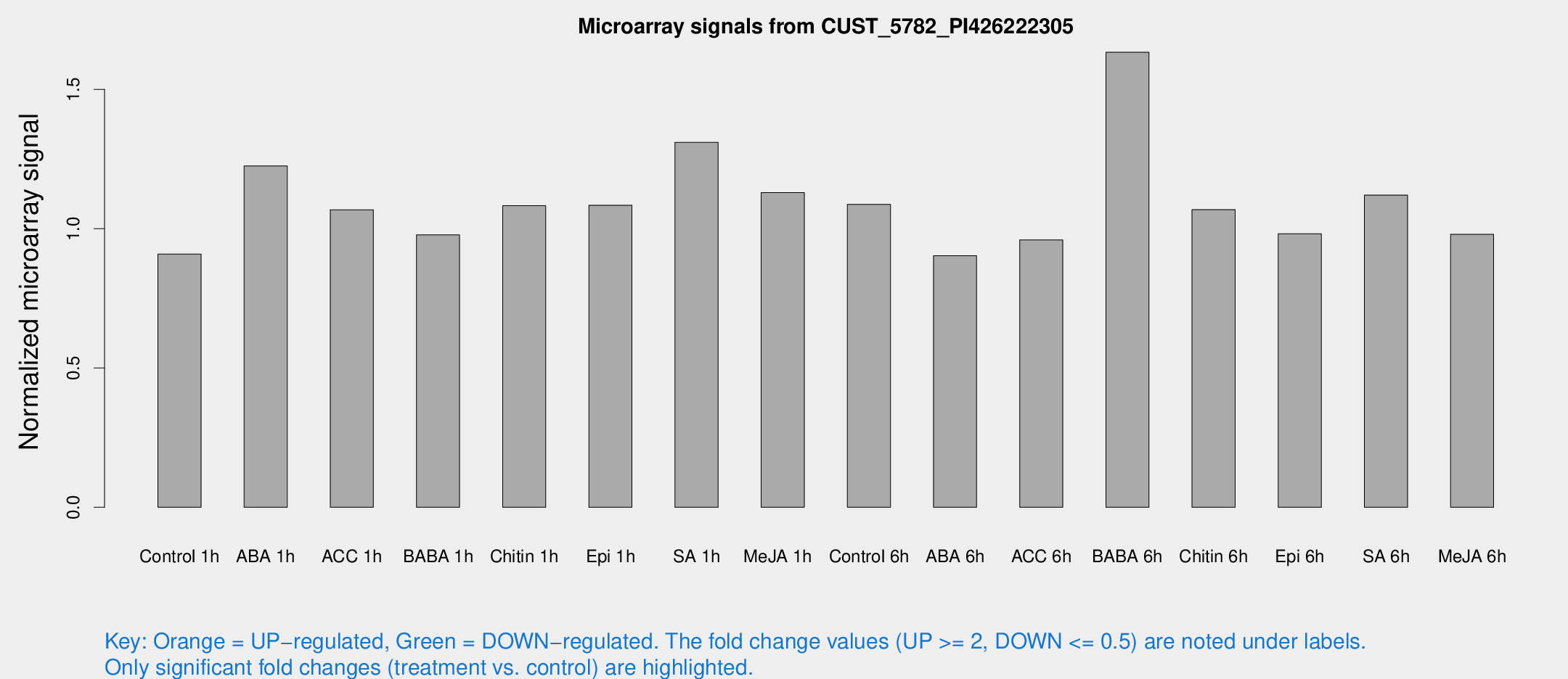
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.51315 | 3.78226 | 0.908735 | 0.526743 |
| ABA 1h | 8.09651 | 3.67557 | 1.22532 | 0.590609 |
| ACC 1h | 8.12963 | 4.88785 | 1.06799 | 0.618782 |
| BABA 1h | 6.74576 | 3.90927 | 0.978128 | 0.566724 |
| Chitin 1h | 7.04365 | 3.79789 | 1.08269 | 0.590489 |
| Epi 1h | 6.7242 | 3.67413 | 1.08423 | 0.593687 |
| SA 1h | 11.1641 | 4.51923 | 1.30979 | 0.590728 |
| Me-JA 1h | 6.60521 | 3.8331 | 1.1292 | 0.65426 |
| Control 6h | 7.84643 | 3.83513 | 1.08733 | 0.538951 |
| ABA 6h | 6.87502 | 3.89768 | 0.902974 | 0.510457 |
| ACC 6h | 8.07841 | 4.80894 | 0.959582 | 0.556286 |
| BABA 6h | 19.676 | 12.5794 | 1.6333 | 1.34332 |
| Chitin 6h | 8.28653 | 4.20835 | 1.0686 | 0.559477 |
| Epi 6h | 7.99489 | 4.6804 | 0.981838 | 0.56937 |
| SA 6h | 8.11316 | 4.07622 | 1.12058 | 0.584778 |
| Me-JA 6h | 7.03448 | 3.81039 | 0.979716 | 0.535749 |
Source Transcript PGSC0003DMT400022917 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G52870.1 | +1 | 4e-82 | 267 | 129/176 (73%) | IQ calmodulin-binding motif family protein | chr3:19593365-19595686 REVERSE LENGTH=456 |
