Probe CUST_5701_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5701_PI426222305 | JHI_St_60k_v1 | DMT400022916 | TGCCATTTGTACTTCTAACATTTGAGTATGAGTTATTCCCTTCCATATATGGTCTCTCAT |
All Microarray Probes Designed to Gene DMG400008882
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5701_PI426222305 | JHI_St_60k_v1 | DMT400022916 | TGCCATTTGTACTTCTAACATTTGAGTATGAGTTATTCCCTTCCATATATGGTCTCTCAT |
| CUST_5782_PI426222305 | JHI_St_60k_v1 | DMT400022917 | ATTGAATACGTGACGATTCCCCCAAAAATAGTGTGTTTTTGGAGGATCCAACACGGGGCT |
| CUST_5629_PI426222305 | JHI_St_60k_v1 | DMT400022914 | TGCCATTTGTACTTCTAACATTTGAGTATGAGTTATTCCCTTCCATATATGGTCTCTCAT |
Microarray Signals from CUST_5701_PI426222305
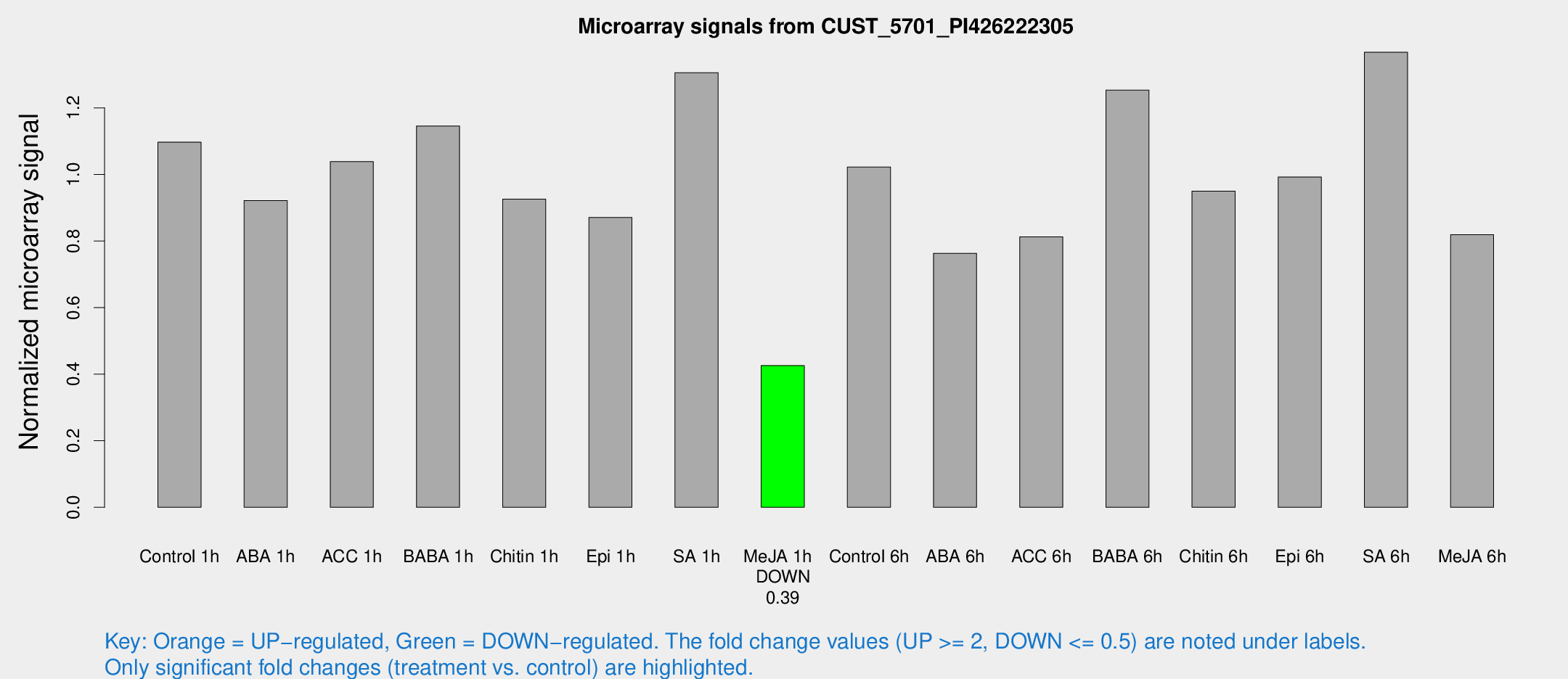
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 453.054 | 64.7754 | 1.09718 | 0.0969561 |
| ABA 1h | 331.009 | 19.4662 | 0.921753 | 0.0541025 |
| ACC 1h | 469.54 | 127.128 | 1.03863 | 0.237983 |
| BABA 1h | 462.099 | 80.4088 | 1.14558 | 0.105495 |
| Chitin 1h | 339.875 | 29.1349 | 0.92569 | 0.0543528 |
| Epi 1h | 311.269 | 42.5129 | 0.870809 | 0.102321 |
| SA 1h | 547.126 | 43.4617 | 1.30587 | 0.145356 |
| Me-JA 1h | 140.941 | 8.96734 | 0.42593 | 0.0348585 |
| Control 6h | 444.306 | 104.292 | 1.02216 | 0.193765 |
| ABA 6h | 337.854 | 52.7074 | 0.763097 | 0.144982 |
| ACC 6h | 378.25 | 22.3423 | 0.812715 | 0.0840766 |
| BABA 6h | 574.04 | 58.7667 | 1.25323 | 0.0995044 |
| Chitin 6h | 410.205 | 24.0832 | 0.949977 | 0.0557133 |
| Epi 6h | 456.561 | 38.8184 | 0.992089 | 0.0643898 |
| SA 6h | 552.422 | 50.8359 | 1.36719 | 0.186089 |
| Me-JA 6h | 356.984 | 103.75 | 0.819136 | 0.167268 |
Source Transcript PGSC0003DMT400022916 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G52870.1 | +1 | 1e-178 | 518 | 269/391 (69%) | IQ calmodulin-binding motif family protein | chr3:19593365-19595686 REVERSE LENGTH=456 |
