Probe CUST_5629_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5629_PI426222305 | JHI_St_60k_v1 | DMT400022914 | TGCCATTTGTACTTCTAACATTTGAGTATGAGTTATTCCCTTCCATATATGGTCTCTCAT |
All Microarray Probes Designed to Gene DMG400008882
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_5701_PI426222305 | JHI_St_60k_v1 | DMT400022916 | TGCCATTTGTACTTCTAACATTTGAGTATGAGTTATTCCCTTCCATATATGGTCTCTCAT |
| CUST_5782_PI426222305 | JHI_St_60k_v1 | DMT400022917 | ATTGAATACGTGACGATTCCCCCAAAAATAGTGTGTTTTTGGAGGATCCAACACGGGGCT |
| CUST_5629_PI426222305 | JHI_St_60k_v1 | DMT400022914 | TGCCATTTGTACTTCTAACATTTGAGTATGAGTTATTCCCTTCCATATATGGTCTCTCAT |
Microarray Signals from CUST_5629_PI426222305
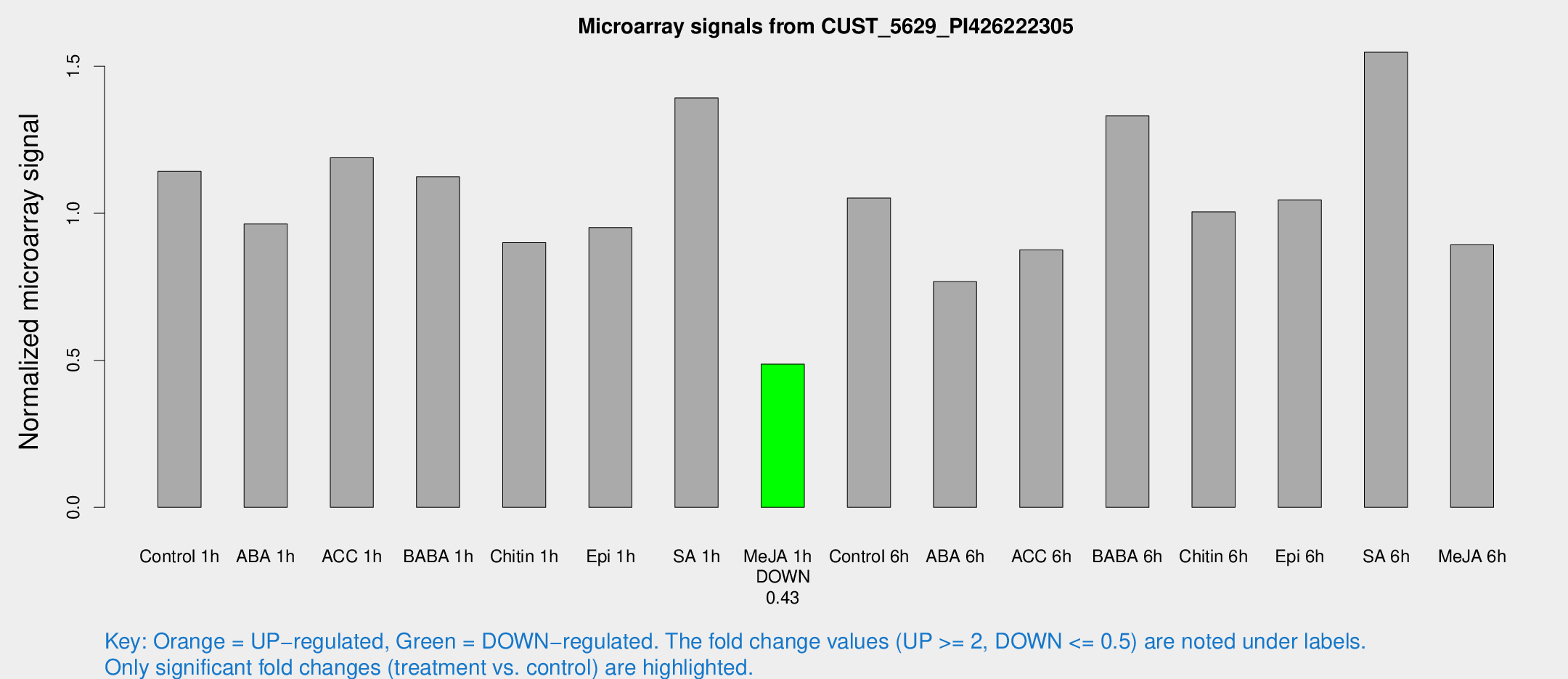
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 485.61 | 81.7284 | 1.14258 | 0.127813 |
| ABA 1h | 362.636 | 60.2443 | 0.963327 | 0.0928726 |
| ACC 1h | 540.053 | 128.011 | 1.18895 | 0.230375 |
| BABA 1h | 460.211 | 71.9653 | 1.12388 | 0.0846977 |
| Chitin 1h | 337.906 | 29.7181 | 0.900336 | 0.0525332 |
| Epi 1h | 347.025 | 45.9535 | 0.951089 | 0.10801 |
| SA 1h | 594.658 | 37.2612 | 1.39253 | 0.115909 |
| Me-JA 1h | 165.084 | 9.99693 | 0.48708 | 0.029409 |
| Control 6h | 460.559 | 96.3955 | 1.05157 | 0.160138 |
| ABA 6h | 342.785 | 39.8688 | 0.767285 | 0.10155 |
| ACC 6h | 422.947 | 57.4647 | 0.875243 | 0.0510423 |
| BABA 6h | 619.75 | 43.0016 | 1.33098 | 0.0771744 |
| Chitin 6h | 446.53 | 40.5675 | 1.00521 | 0.102594 |
| Epi 6h | 487.96 | 28.3824 | 1.0452 | 0.072114 |
| SA 6h | 650.834 | 99.995 | 1.54772 | 0.378211 |
| Me-JA 6h | 389.417 | 93.7509 | 0.892583 | 0.140359 |
Source Transcript PGSC0003DMT400022914 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G52870.1 | +1 | 2e-76 | 257 | 147/247 (60%) | IQ calmodulin-binding motif family protein | chr3:19593365-19595686 REVERSE LENGTH=456 |
