Probe CUST_51696_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51696_PI426222305 | JHI_St_60k_v1 | DMT400017094 | GAGTGGTTGTTCCTTTCTAGTGACTGTCTTTAGCATTTTGTCTTGGTTTCTTCTCTCCTG |
All Microarray Probes Designed to Gene DMG400006672
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51690_PI426222305 | JHI_St_60k_v1 | DMT400017092 | TGTCATCTAGTTCCTCTTTCCCATAAACATTGTCACCTAGTTTTTCCTACAATCTAAAGA |
| CUST_51695_PI426222305 | JHI_St_60k_v1 | DMT400017088 | AATATAAATGCAGTTCAAGTACTATTATACGCCTAAGCATGTCTGCGCTTGGGGCTGGGG |
| CUST_51692_PI426222305 | JHI_St_60k_v1 | DMT400017093 | TCACTAGTACCTATCAAGAGATTGCAGGAACGTAAGTCATCTTAATATGGCCCTGGTTTT |
| CUST_51694_PI426222305 | JHI_St_60k_v1 | DMT400017087 | GAGTTGGTAAAATCTGTTCCTCTAGCAATTGATGACAAGCACTGAGCAATGATGGTGCAA |
| CUST_51696_PI426222305 | JHI_St_60k_v1 | DMT400017094 | GAGTGGTTGTTCCTTTCTAGTGACTGTCTTTAGCATTTTGTCTTGGTTTCTTCTCTCCTG |
Microarray Signals from CUST_51696_PI426222305
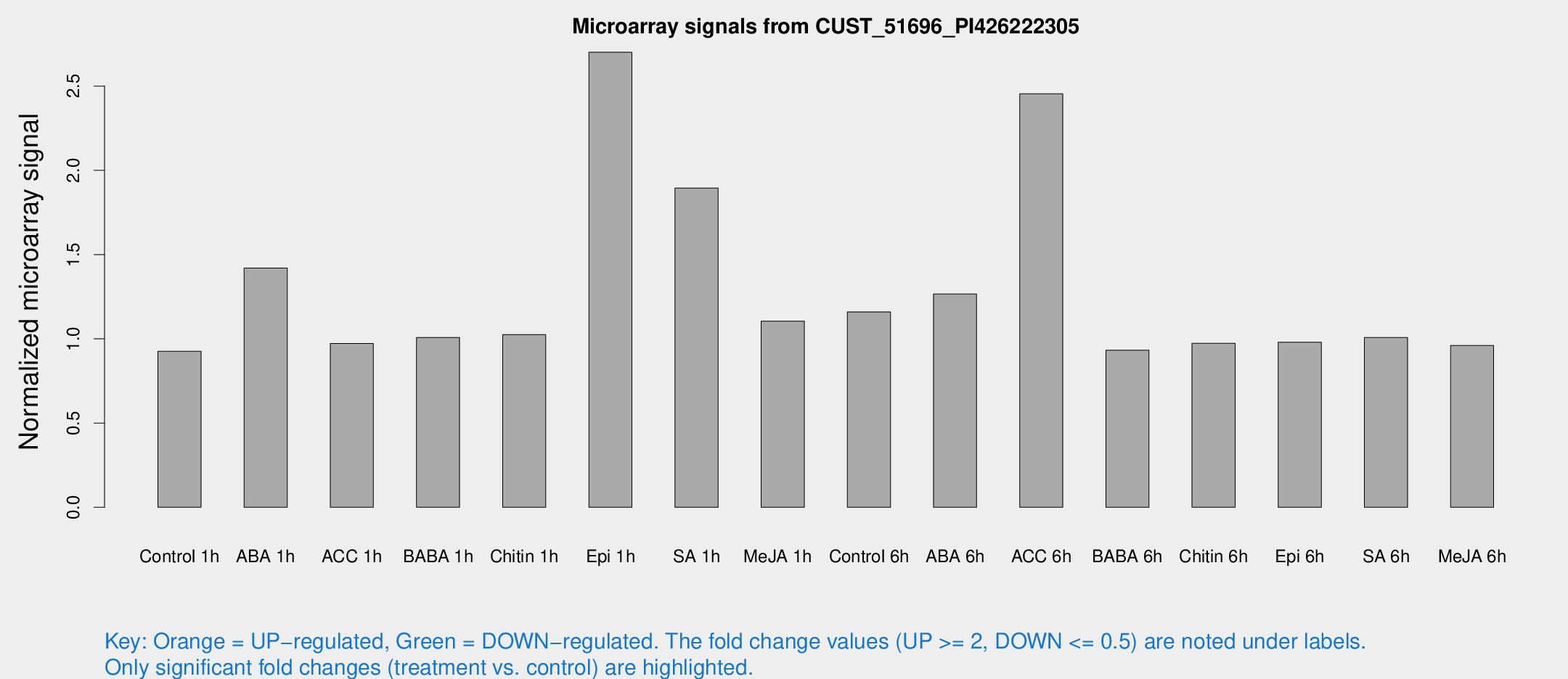
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.50379 | 3.65466 | 0.925971 | 0.521491 |
| ABA 1h | 9.35508 | 3.54261 | 1.41997 | 0.603227 |
| ACC 1h | 7.02784 | 4.08686 | 0.971889 | 0.562972 |
| BABA 1h | 6.81745 | 3.94849 | 1.00815 | 0.583775 |
| Chitin 1h | 6.48209 | 3.76272 | 1.02516 | 0.59366 |
| Epi 1h | 43.5436 | 37.4418 | 2.70105 | 6.93182 |
| SA 1h | 25.8108 | 19.5349 | 1.89507 | 2.15039 |
| Me-JA 1h | 6.34543 | 3.68517 | 1.1049 | 0.639761 |
| Control 6h | 8.4414 | 3.93904 | 1.15945 | 0.581648 |
| ABA 6h | 9.8799 | 3.90518 | 1.26609 | 0.551598 |
| ACC 6h | 24.5112 | 11.6949 | 2.45405 | 1.68299 |
| BABA 6h | 7.34776 | 4.27445 | 0.93199 | 0.539875 |
| Chitin 6h | 7.26548 | 4.20746 | 0.972937 | 0.563354 |
| Epi 6h | 7.81595 | 4.5692 | 0.979524 | 0.567866 |
| SA 6h | 6.98982 | 4.04791 | 1.00739 | 0.583361 |
| Me-JA 6h | 6.72082 | 3.81311 | 0.960271 | 0.54502 |
Source Transcript PGSC0003DMT400017094 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G49190.1 | +3 | 5e-36 | 139 | 64/75 (85%) | sucrose synthase 2 | chr5:19943369-19947189 REVERSE LENGTH=807 |
