Probe CUST_51695_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51695_PI426222305 | JHI_St_60k_v1 | DMT400017088 | AATATAAATGCAGTTCAAGTACTATTATACGCCTAAGCATGTCTGCGCTTGGGGCTGGGG |
All Microarray Probes Designed to Gene DMG400006672
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51690_PI426222305 | JHI_St_60k_v1 | DMT400017092 | TGTCATCTAGTTCCTCTTTCCCATAAACATTGTCACCTAGTTTTTCCTACAATCTAAAGA |
| CUST_51695_PI426222305 | JHI_St_60k_v1 | DMT400017088 | AATATAAATGCAGTTCAAGTACTATTATACGCCTAAGCATGTCTGCGCTTGGGGCTGGGG |
| CUST_51692_PI426222305 | JHI_St_60k_v1 | DMT400017093 | TCACTAGTACCTATCAAGAGATTGCAGGAACGTAAGTCATCTTAATATGGCCCTGGTTTT |
| CUST_51694_PI426222305 | JHI_St_60k_v1 | DMT400017087 | GAGTTGGTAAAATCTGTTCCTCTAGCAATTGATGACAAGCACTGAGCAATGATGGTGCAA |
| CUST_51696_PI426222305 | JHI_St_60k_v1 | DMT400017094 | GAGTGGTTGTTCCTTTCTAGTGACTGTCTTTAGCATTTTGTCTTGGTTTCTTCTCTCCTG |
Microarray Signals from CUST_51695_PI426222305
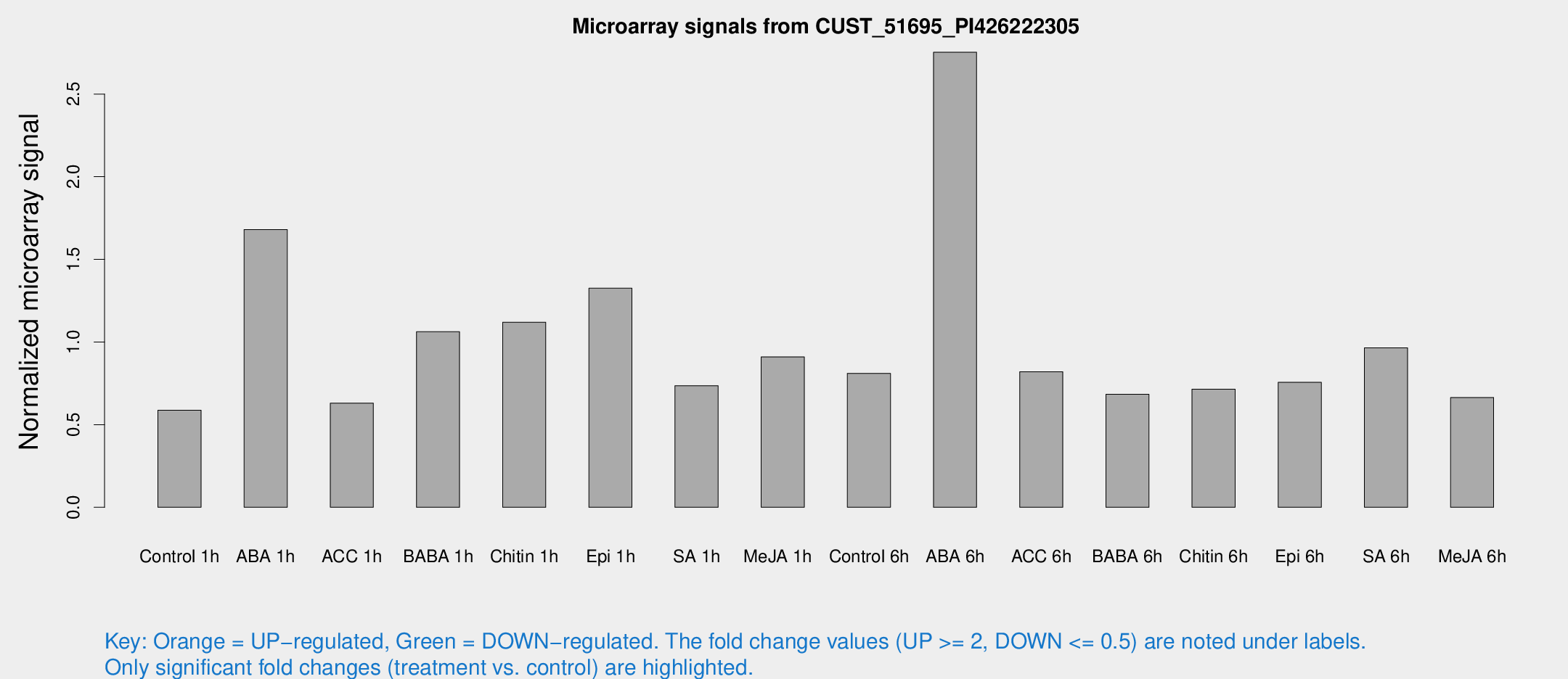
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.22307 | 3.08457 | 0.587495 | 0.276271 |
| ABA 1h | 19.1051 | 3.17437 | 1.68056 | 0.293399 |
| ACC 1h | 8.45821 | 3.37884 | 0.630311 | 0.271101 |
| BABA 1h | 13.0017 | 3.30909 | 1.06231 | 0.273773 |
| Chitin 1h | 12.9792 | 3.20172 | 1.11967 | 0.282989 |
| Epi 1h | 14.8437 | 3.15615 | 1.32527 | 0.30146 |
| SA 1h | 10.1989 | 3.11371 | 0.735608 | 0.26148 |
| Me-JA 1h | 9.83501 | 3.218 | 0.909876 | 0.333924 |
| Control 6h | 10.7759 | 3.32352 | 0.810532 | 0.278467 |
| ABA 6h | 38.6626 | 8.51245 | 2.75307 | 0.440147 |
| ACC 6h | 12.4835 | 3.9548 | 0.820102 | 0.304341 |
| BABA 6h | 10.3188 | 3.68442 | 0.683679 | 0.274538 |
| Chitin 6h | 10.4796 | 3.60179 | 0.7145 | 0.298418 |
| Epi 6h | 11.2241 | 3.757 | 0.75596 | 0.277758 |
| SA 6h | 13.3722 | 3.70168 | 0.96424 | 0.336353 |
| Me-JA 6h | 9.68571 | 3.93362 | 0.664081 | 0.351473 |
Source Transcript PGSC0003DMT400017088 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G02280.1 | +1 | 0.0 | 629 | 309/394 (78%) | sucrose synthase 3 | chr4:995166-998719 FORWARD LENGTH=809 |
