Probe CUST_51694_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51694_PI426222305 | JHI_St_60k_v1 | DMT400017087 | GAGTTGGTAAAATCTGTTCCTCTAGCAATTGATGACAAGCACTGAGCAATGATGGTGCAA |
All Microarray Probes Designed to Gene DMG400006672
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_51690_PI426222305 | JHI_St_60k_v1 | DMT400017092 | TGTCATCTAGTTCCTCTTTCCCATAAACATTGTCACCTAGTTTTTCCTACAATCTAAAGA |
| CUST_51695_PI426222305 | JHI_St_60k_v1 | DMT400017088 | AATATAAATGCAGTTCAAGTACTATTATACGCCTAAGCATGTCTGCGCTTGGGGCTGGGG |
| CUST_51692_PI426222305 | JHI_St_60k_v1 | DMT400017093 | TCACTAGTACCTATCAAGAGATTGCAGGAACGTAAGTCATCTTAATATGGCCCTGGTTTT |
| CUST_51694_PI426222305 | JHI_St_60k_v1 | DMT400017087 | GAGTTGGTAAAATCTGTTCCTCTAGCAATTGATGACAAGCACTGAGCAATGATGGTGCAA |
| CUST_51696_PI426222305 | JHI_St_60k_v1 | DMT400017094 | GAGTGGTTGTTCCTTTCTAGTGACTGTCTTTAGCATTTTGTCTTGGTTTCTTCTCTCCTG |
Microarray Signals from CUST_51694_PI426222305
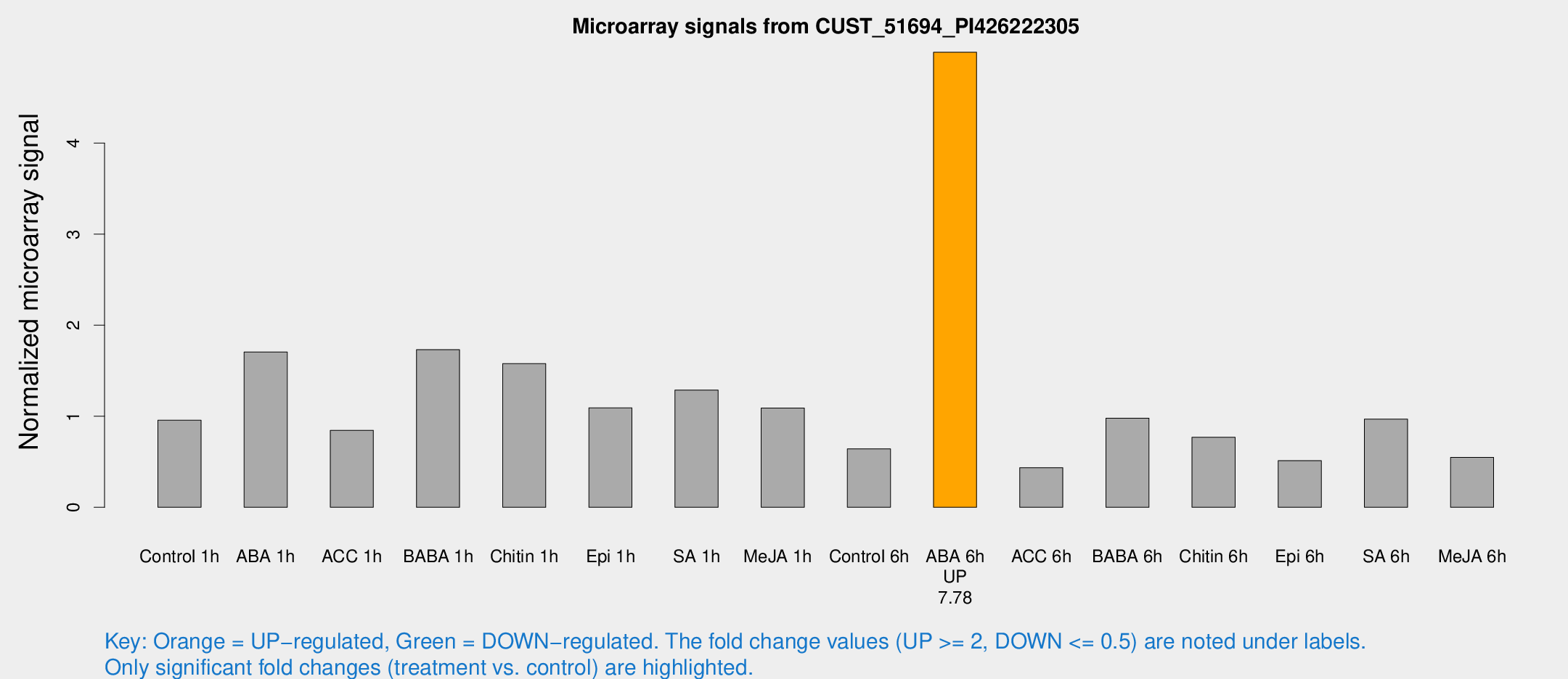
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 164.821 | 18.1422 | 0.956877 | 0.0678958 |
| ABA 1h | 269.949 | 58.0089 | 1.70666 | 0.348115 |
| ACC 1h | 159.152 | 38.3178 | 0.845772 | 0.184615 |
| BABA 1h | 286.641 | 29.669 | 1.73091 | 0.10206 |
| Chitin 1h | 260.139 | 71.2922 | 1.57808 | 0.317949 |
| Epi 1h | 163.375 | 21.5073 | 1.09233 | 0.146311 |
| SA 1h | 231.006 | 35.9104 | 1.28776 | 0.142444 |
| Me-JA 1h | 152.427 | 13.5449 | 1.08954 | 0.0670806 |
| Control 6h | 122.404 | 34.6273 | 0.642496 | 0.180351 |
| ABA 6h | 906.388 | 53.2281 | 4.99875 | 0.289225 |
| ACC 6h | 89.0199 | 19.2076 | 0.4359 | 0.0403918 |
| BABA 6h | 186.51 | 11.3914 | 0.979342 | 0.0673912 |
| Chitin 6h | 140.008 | 11.0373 | 0.768891 | 0.0528408 |
| Epi 6h | 99.6065 | 12.0449 | 0.512224 | 0.0561816 |
| SA 6h | 166.998 | 25.3537 | 0.968095 | 0.0600928 |
| Me-JA 6h | 93.7411 | 9.33024 | 0.548002 | 0.0372304 |
Source Transcript PGSC0003DMT400017087 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G02280.1 | +1 | 0.0 | 1318 | 634/809 (78%) | sucrose synthase 3 | chr4:995166-998719 FORWARD LENGTH=809 |
