Probe CUST_49034_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49034_PI426222305 | JHI_St_60k_v1 | DMT400021899 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
All Microarray Probes Designed to Gene DMG400008496
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49003_PI426222305 | JHI_St_60k_v1 | DMT400021901 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
| CUST_49034_PI426222305 | JHI_St_60k_v1 | DMT400021899 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
| CUST_49001_PI426222305 | JHI_St_60k_v1 | DMT400021900 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
Microarray Signals from CUST_49034_PI426222305
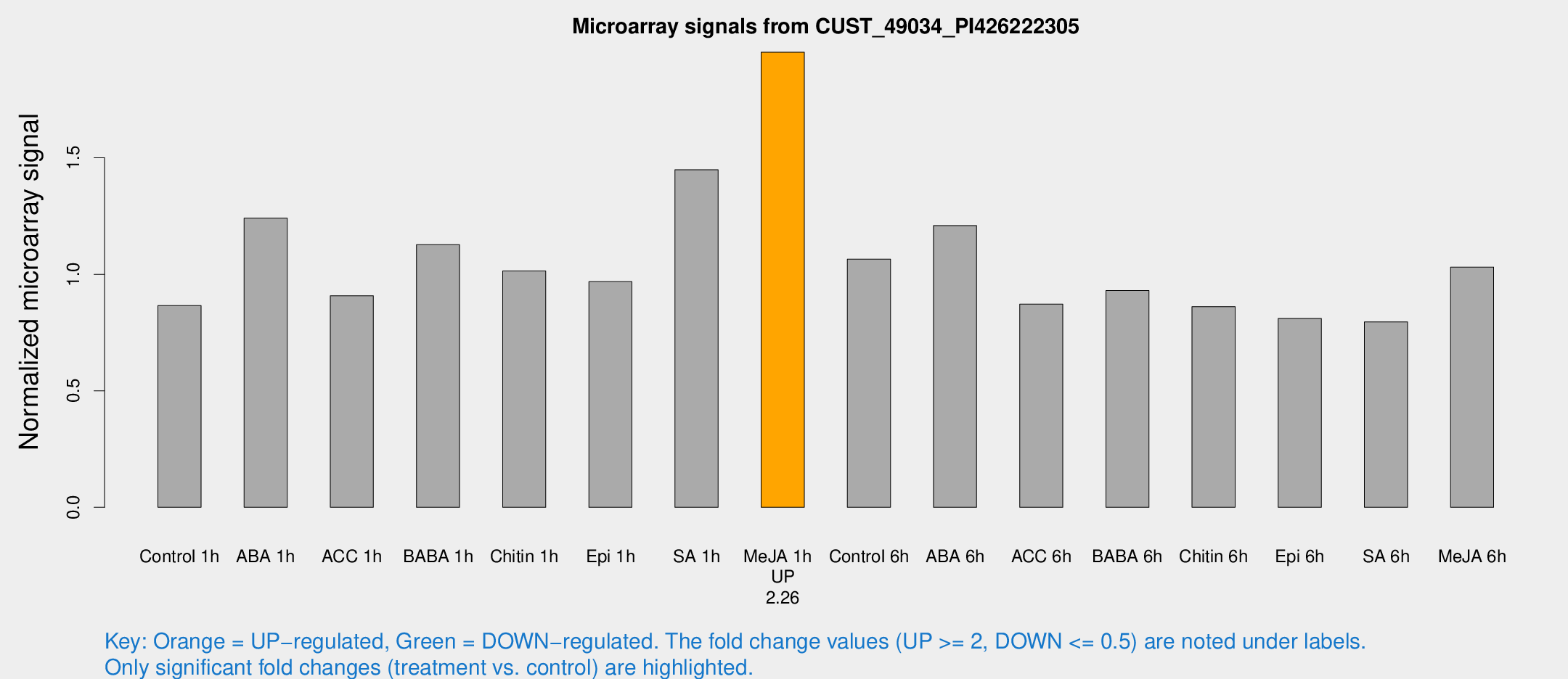
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 323.33 | 40.7817 | 0.865858 | 0.056889 |
| ABA 1h | 431.459 | 111.085 | 1.24072 | 0.238786 |
| ACC 1h | 366.123 | 95.9914 | 0.90797 | 0.194961 |
| BABA 1h | 420.436 | 90.4339 | 1.1272 | 0.157601 |
| Chitin 1h | 345.289 | 54.1896 | 1.01456 | 0.117071 |
| Epi 1h | 310.33 | 24.6686 | 0.968202 | 0.0600596 |
| SA 1h | 576.255 | 135.487 | 1.4487 | 0.323094 |
| Me-JA 1h | 593.708 | 66.3207 | 1.95323 | 0.113281 |
| Control 6h | 421.545 | 101.94 | 1.06515 | 0.21005 |
| ABA 6h | 480.407 | 61.6337 | 1.20934 | 0.0870022 |
| ACC 6h | 369.901 | 26.2154 | 0.871831 | 0.0874183 |
| BABA 6h | 384.112 | 22.5488 | 0.930116 | 0.0544536 |
| Chitin 6h | 337.538 | 19.8359 | 0.86145 | 0.0506018 |
| Epi 6h | 337.09 | 19.8807 | 0.81048 | 0.0760605 |
| SA 6h | 316.396 | 82.6357 | 0.79571 | 0.16264 |
| Me-JA 6h | 389.724 | 64.4285 | 1.03076 | 0.10897 |
Source Transcript PGSC0003DMT400021899 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G38280.1 | +1 | 9e-133 | 415 | 196/215 (91%) | AMP deaminase, putative / myoadenylate deaminase, putative | chr2:16033767-16038793 REVERSE LENGTH=839 |
