Probe CUST_49001_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49001_PI426222305 | JHI_St_60k_v1 | DMT400021900 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
All Microarray Probes Designed to Gene DMG400008496
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49003_PI426222305 | JHI_St_60k_v1 | DMT400021901 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
| CUST_49034_PI426222305 | JHI_St_60k_v1 | DMT400021899 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
| CUST_49001_PI426222305 | JHI_St_60k_v1 | DMT400021900 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
Microarray Signals from CUST_49001_PI426222305
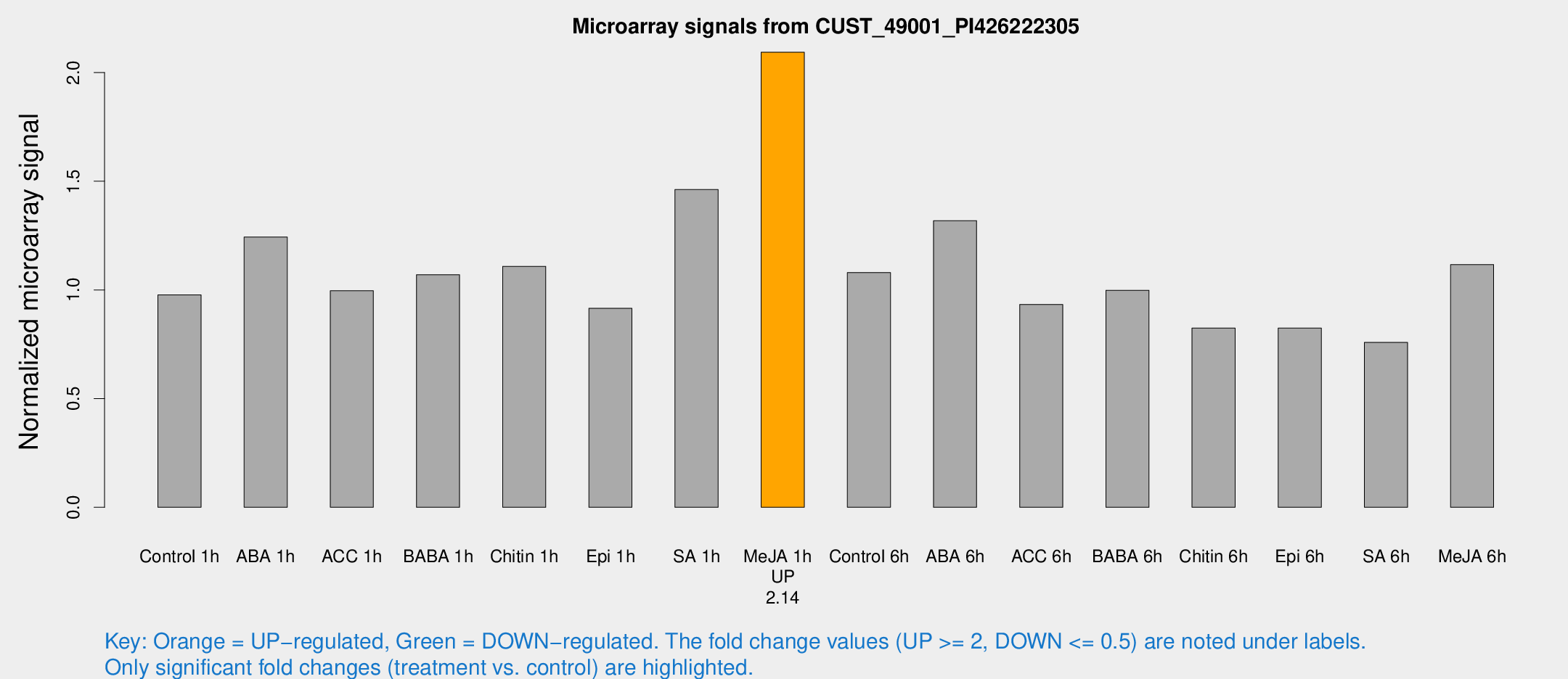
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 351.038 | 23.9279 | 0.976895 | 0.0573583 |
| ABA 1h | 414.884 | 95.539 | 1.24335 | 0.203184 |
| ACC 1h | 379.586 | 75.8892 | 0.996136 | 0.174951 |
| BABA 1h | 375.63 | 47.7763 | 1.06999 | 0.0628268 |
| Chitin 1h | 362.195 | 44.569 | 1.10847 | 0.114365 |
| Epi 1h | 285.403 | 23.7865 | 0.915354 | 0.0554387 |
| SA 1h | 576.409 | 161.069 | 1.46175 | 0.407257 |
| Me-JA 1h | 618.719 | 64.752 | 2.09353 | 0.121579 |
| Control 6h | 428.373 | 119.943 | 1.0799 | 0.281805 |
| ABA 6h | 505.845 | 48.2219 | 1.31848 | 0.082195 |
| ACC 6h | 384.796 | 26.5135 | 0.933077 | 0.0825106 |
| BABA 6h | 401.977 | 29.7567 | 0.998399 | 0.0586266 |
| Chitin 6h | 314.166 | 18.6581 | 0.824829 | 0.0489761 |
| Epi 6h | 333.244 | 19.8706 | 0.824436 | 0.0572356 |
| SA 6h | 287.267 | 67.0385 | 0.75843 | 0.121255 |
| Me-JA 6h | 415.297 | 82.0493 | 1.11644 | 0.144726 |
Source Transcript PGSC0003DMT400021900 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G38280.1 | +1 | 2e-172 | 523 | 252/273 (92%) | AMP deaminase, putative / myoadenylate deaminase, putative | chr2:16033767-16038793 REVERSE LENGTH=839 |
