Probe CUST_49003_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49003_PI426222305 | JHI_St_60k_v1 | DMT400021901 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
All Microarray Probes Designed to Gene DMG400008496
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_49003_PI426222305 | JHI_St_60k_v1 | DMT400021901 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
| CUST_49034_PI426222305 | JHI_St_60k_v1 | DMT400021899 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
| CUST_49001_PI426222305 | JHI_St_60k_v1 | DMT400021900 | GAACAAGGTAGGGTTGGATTGTGACTAGAACTATTTTTTGATGTTGGTTTCTCTTTTTTG |
Microarray Signals from CUST_49003_PI426222305
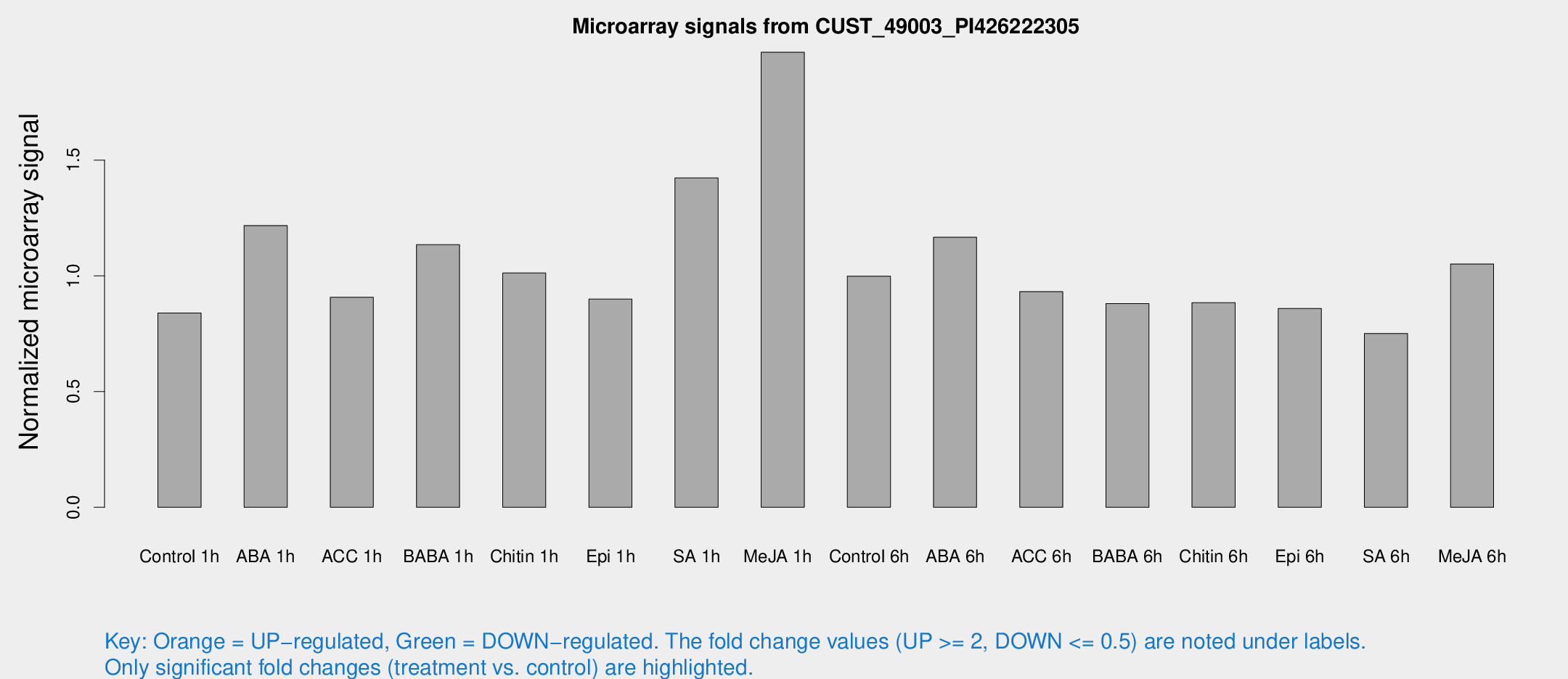
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 309.795 | 34.3436 | 0.839335 | 0.0678067 |
| ABA 1h | 410.818 | 90.9901 | 1.21742 | 0.17725 |
| ACC 1h | 350.658 | 64.2541 | 0.907497 | 0.136054 |
| BABA 1h | 405.901 | 51.4493 | 1.13464 | 0.066395 |
| Chitin 1h | 338.257 | 43.4578 | 1.01257 | 0.0852639 |
| Epi 1h | 289.315 | 37.4003 | 0.899582 | 0.115313 |
| SA 1h | 554.944 | 115.971 | 1.4232 | 0.29434 |
| Me-JA 1h | 598.959 | 87.0014 | 1.9661 | 0.14245 |
| Control 6h | 389.655 | 90.3293 | 0.99879 | 0.183463 |
| ABA 6h | 457.249 | 48.2036 | 1.16687 | 0.0681451 |
| ACC 6h | 394.086 | 39.9289 | 0.931519 | 0.0868318 |
| BABA 6h | 362.029 | 30.7028 | 0.880525 | 0.0518417 |
| Chitin 6h | 343.376 | 20.2945 | 0.88406 | 0.0522289 |
| Epi 6h | 354.692 | 22.2975 | 0.8591 | 0.0987101 |
| SA 6h | 304.308 | 87.6461 | 0.750984 | 0.194771 |
| Me-JA 6h | 396.002 | 70.7496 | 1.05123 | 0.12154 |
Source Transcript PGSC0003DMT400021901 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G38280.1 | +2 | 0.0 | 1317 | 667/851 (78%) | AMP deaminase, putative / myoadenylate deaminase, putative | chr2:16033767-16038793 REVERSE LENGTH=839 |
