Probe CUST_46856_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46856_PI426222305 | JHI_St_60k_v1 | DMT400081726 | GAGAAACGAAAAGTGACACATCAAACTTGAGGCATCTCATAGGAAATCTCCTTACCAAGA |
All Microarray Probes Designed to Gene DMG400032094
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46853_PI426222305 | JHI_St_60k_v1 | DMT400081727 | TCGCGATCCATTTTGCATAGAAAAGAACAGGAGAGGCAGATGCCAGGTGATGAATTGTTT |
| CUST_46856_PI426222305 | JHI_St_60k_v1 | DMT400081726 | GAGAAACGAAAAGTGACACATCAAACTTGAGGCATCTCATAGGAAATCTCCTTACCAAGA |
| CUST_46867_PI426222305 | JHI_St_60k_v1 | DMT400081725 | GAGAAACGAAAAGTGACACATCAAACTTGAGGCATCTCATAGGAAATCTCCTTACCAAGA |
Microarray Signals from CUST_46856_PI426222305
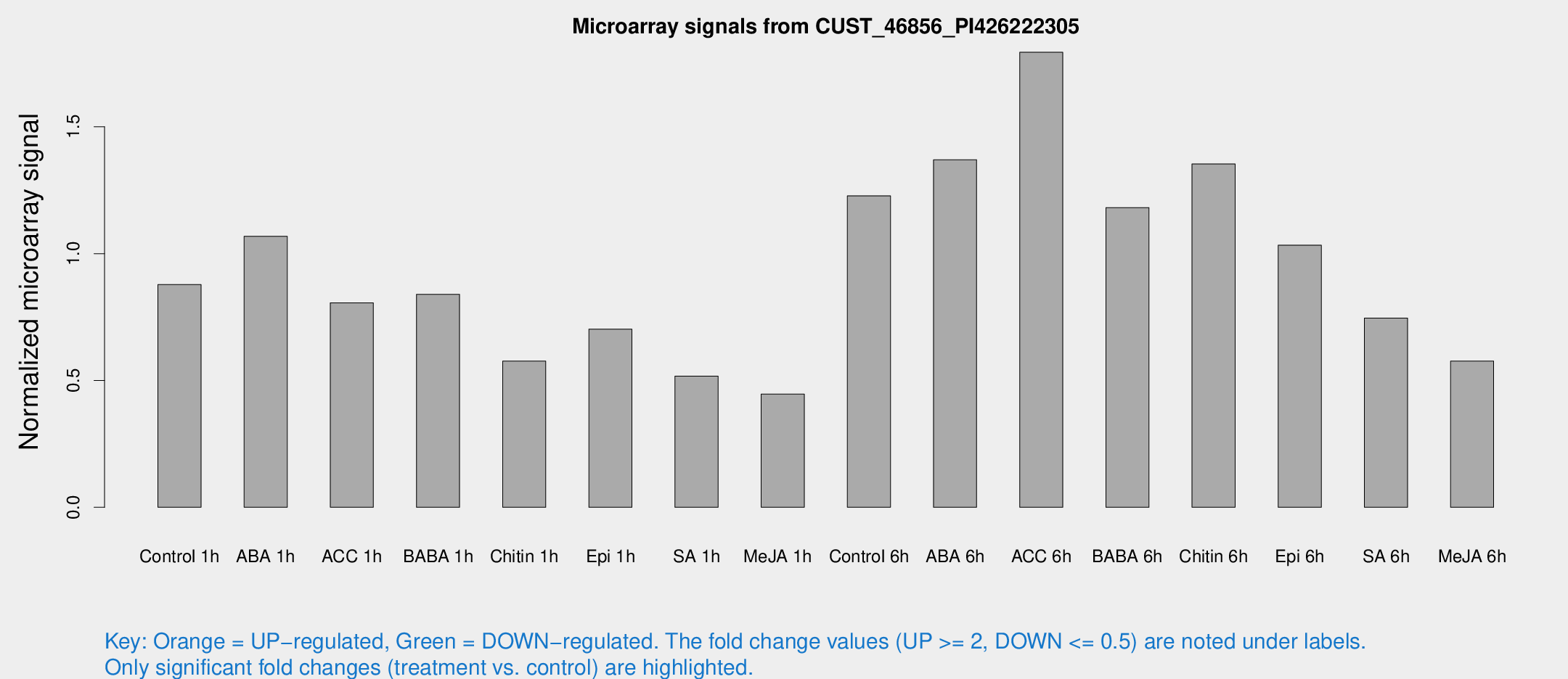
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 15.3856 | 4.8806 | 0.878264 | 0.23371 |
| ABA 1h | 17.5955 | 6.42412 | 1.06835 | 0.548011 |
| ACC 1h | 14.8396 | 5.01102 | 0.806229 | 0.320532 |
| BABA 1h | 14.4057 | 4.54903 | 0.839525 | 0.338894 |
| Chitin 1h | 8.75443 | 3.36958 | 0.576429 | 0.242986 |
| Epi 1h | 10.9151 | 3.89598 | 0.702285 | 0.278787 |
| SA 1h | 8.66415 | 3.32479 | 0.516797 | 0.213484 |
| Me-JA 1h | 5.85209 | 3.42288 | 0.446032 | 0.258675 |
| Control 6h | 19.8766 | 3.73285 | 1.22794 | 0.241174 |
| ABA 6h | 25.1514 | 6.25448 | 1.36997 | 0.453152 |
| ACC 6h | 33.3174 | 4.64311 | 1.79392 | 0.243341 |
| BABA 6h | 21.4487 | 4.06211 | 1.18156 | 0.233178 |
| Chitin 6h | 23.2159 | 4.11926 | 1.35337 | 0.245226 |
| Epi 6h | 18.9401 | 4.17054 | 1.03348 | 0.226438 |
| SA 6h | 14.3058 | 6.56623 | 0.745573 | 0.520761 |
| Me-JA 6h | 10.1628 | 3.52969 | 0.576441 | 0.250561 |
Source Transcript PGSC0003DMT400081726 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G18860.1 | +2 | 0.0 | 672 | 346/557 (62%) | inosine-uridine preferring nucleoside hydrolase family protein | chr5:6291023-6295581 FORWARD LENGTH=890 |
