Probe CUST_46867_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46867_PI426222305 | JHI_St_60k_v1 | DMT400081725 | GAGAAACGAAAAGTGACACATCAAACTTGAGGCATCTCATAGGAAATCTCCTTACCAAGA |
All Microarray Probes Designed to Gene DMG400032094
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46853_PI426222305 | JHI_St_60k_v1 | DMT400081727 | TCGCGATCCATTTTGCATAGAAAAGAACAGGAGAGGCAGATGCCAGGTGATGAATTGTTT |
| CUST_46856_PI426222305 | JHI_St_60k_v1 | DMT400081726 | GAGAAACGAAAAGTGACACATCAAACTTGAGGCATCTCATAGGAAATCTCCTTACCAAGA |
| CUST_46867_PI426222305 | JHI_St_60k_v1 | DMT400081725 | GAGAAACGAAAAGTGACACATCAAACTTGAGGCATCTCATAGGAAATCTCCTTACCAAGA |
Microarray Signals from CUST_46867_PI426222305
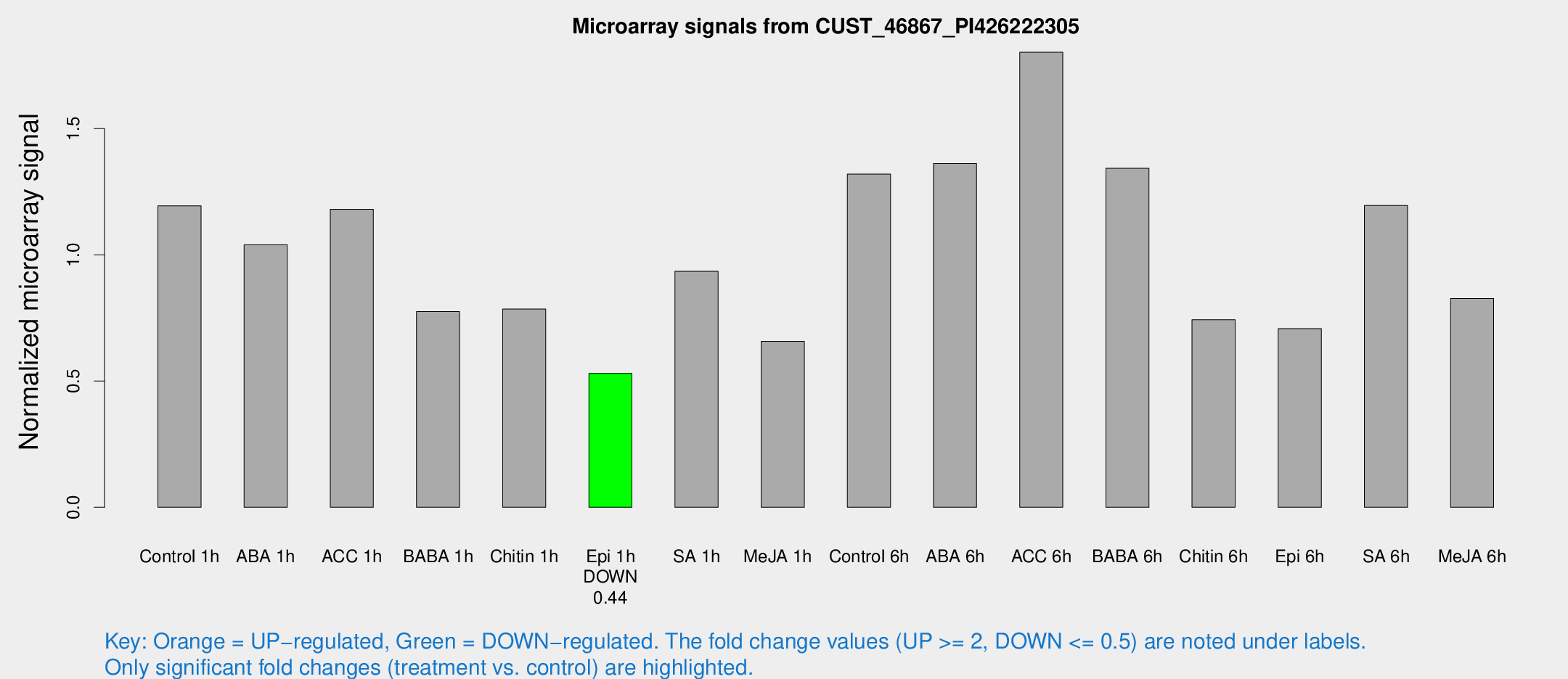
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 18.8182 | 3.79053 | 1.19437 | 0.252004 |
| ABA 1h | 14.6369 | 3.60646 | 1.03989 | 0.26591 |
| ACC 1h | 18.9048 | 4.04308 | 1.18088 | 0.258785 |
| BABA 1h | 12.5865 | 3.84834 | 0.775179 | 0.289326 |
| Chitin 1h | 13.1676 | 5.67699 | 0.785361 | 0.360807 |
| Epi 1h | 7.1712 | 3.54579 | 0.530741 | 0.26578 |
| SA 1h | 14.955 | 3.71049 | 0.934597 | 0.234054 |
| Me-JA 1h | 8.9076 | 3.66527 | 0.657282 | 0.314055 |
| Control 6h | 33.8417 | 22.162 | 1.31968 | 1.2554 |
| ABA 6h | 23.1384 | 3.98688 | 1.36186 | 0.326841 |
| ACC 6h | 34.4825 | 8.18811 | 1.80265 | 0.483235 |
| BABA 6h | 23.7762 | 4.11723 | 1.34314 | 0.248003 |
| Chitin 6h | 13.1212 | 4.14876 | 0.743336 | 0.280264 |
| Epi 6h | 12.6422 | 4.45679 | 0.707481 | 0.270039 |
| SA 6h | 20.3401 | 6.71789 | 1.19548 | 0.596132 |
| Me-JA 6h | 12.9057 | 3.75227 | 0.826761 | 0.246403 |
Source Transcript PGSC0003DMT400081725 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G18860.1 | +2 | 0.0 | 1130 | 565/857 (66%) | inosine-uridine preferring nucleoside hydrolase family protein | chr5:6291023-6295581 FORWARD LENGTH=890 |
