Probe CUST_46853_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46853_PI426222305 | JHI_St_60k_v1 | DMT400081727 | TCGCGATCCATTTTGCATAGAAAAGAACAGGAGAGGCAGATGCCAGGTGATGAATTGTTT |
All Microarray Probes Designed to Gene DMG400032094
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_46853_PI426222305 | JHI_St_60k_v1 | DMT400081727 | TCGCGATCCATTTTGCATAGAAAAGAACAGGAGAGGCAGATGCCAGGTGATGAATTGTTT |
| CUST_46856_PI426222305 | JHI_St_60k_v1 | DMT400081726 | GAGAAACGAAAAGTGACACATCAAACTTGAGGCATCTCATAGGAAATCTCCTTACCAAGA |
| CUST_46867_PI426222305 | JHI_St_60k_v1 | DMT400081725 | GAGAAACGAAAAGTGACACATCAAACTTGAGGCATCTCATAGGAAATCTCCTTACCAAGA |
Microarray Signals from CUST_46853_PI426222305
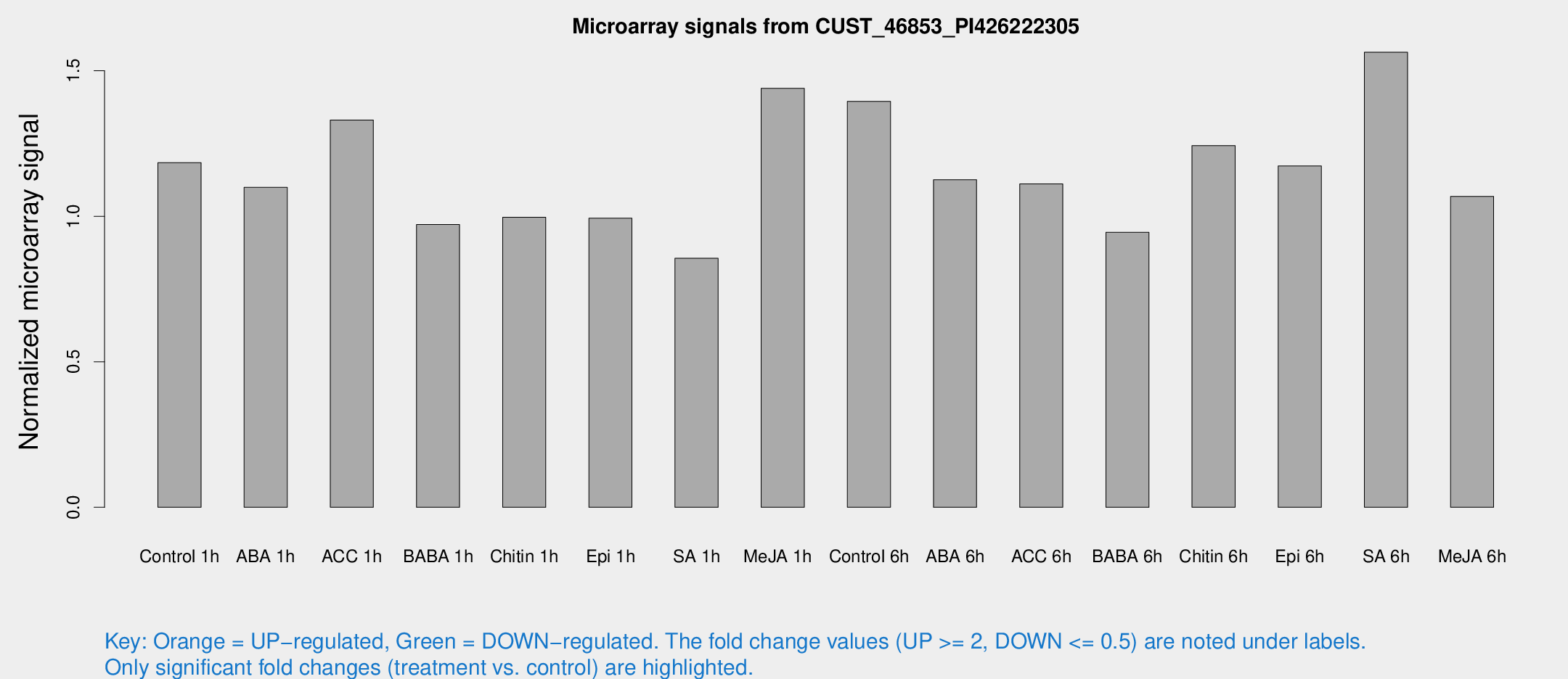
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.32628 | 3.62151 | 1.18422 | 0.563158 |
| ABA 1h | 7.01965 | 3.5254 | 1.09913 | 0.573207 |
| ACC 1h | 9.95778 | 4.01926 | 1.33037 | 0.574176 |
| BABA 1h | 6.62974 | 3.85491 | 0.971562 | 0.562992 |
| Chitin 1h | 6.36002 | 3.71147 | 0.996329 | 0.577923 |
| Epi 1h | 6.08651 | 3.51832 | 0.993327 | 0.57147 |
| SA 1h | 6.21355 | 3.60527 | 0.855603 | 0.495466 |
| Me-JA 1h | 8.70159 | 3.9987 | 1.4394 | 0.717335 |
| Control 6h | 11.8298 | 5.27433 | 1.39475 | 0.648839 |
| ABA 6h | 8.9625 | 4.02885 | 1.12545 | 0.5589 |
| ACC 6h | 9.26988 | 4.60417 | 1.11108 | 0.556606 |
| BABA 6h | 7.50499 | 4.38131 | 0.944928 | 0.548607 |
| Chitin 6h | 9.72358 | 4.36951 | 1.24305 | 0.612134 |
| Epi 6h | 9.99904 | 4.45284 | 1.17276 | 0.557868 |
| SA 6h | 13.2694 | 6.13619 | 1.56365 | 1.0845 |
| Me-JA 6h | 7.55815 | 3.95056 | 1.06819 | 0.566431 |
Source Transcript PGSC0003DMT400081727 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G18860.1 | +2 | 7e-166 | 468 | 221/305 (72%) | inosine-uridine preferring nucleoside hydrolase family protein | chr5:6291023-6295581 FORWARD LENGTH=890 |
