Probe CUST_43675_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43675_PI426222305 | JHI_St_60k_v1 | DMT400004867 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
All Microarray Probes Designed to Gene DMG400001934
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43685_PI426222305 | JHI_St_60k_v1 | DMT400004865 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43704_PI426222305 | JHI_St_60k_v1 | DMT400004864 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43690_PI426222305 | JHI_St_60k_v1 | DMT400004866 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43675_PI426222305 | JHI_St_60k_v1 | DMT400004867 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
Microarray Signals from CUST_43675_PI426222305
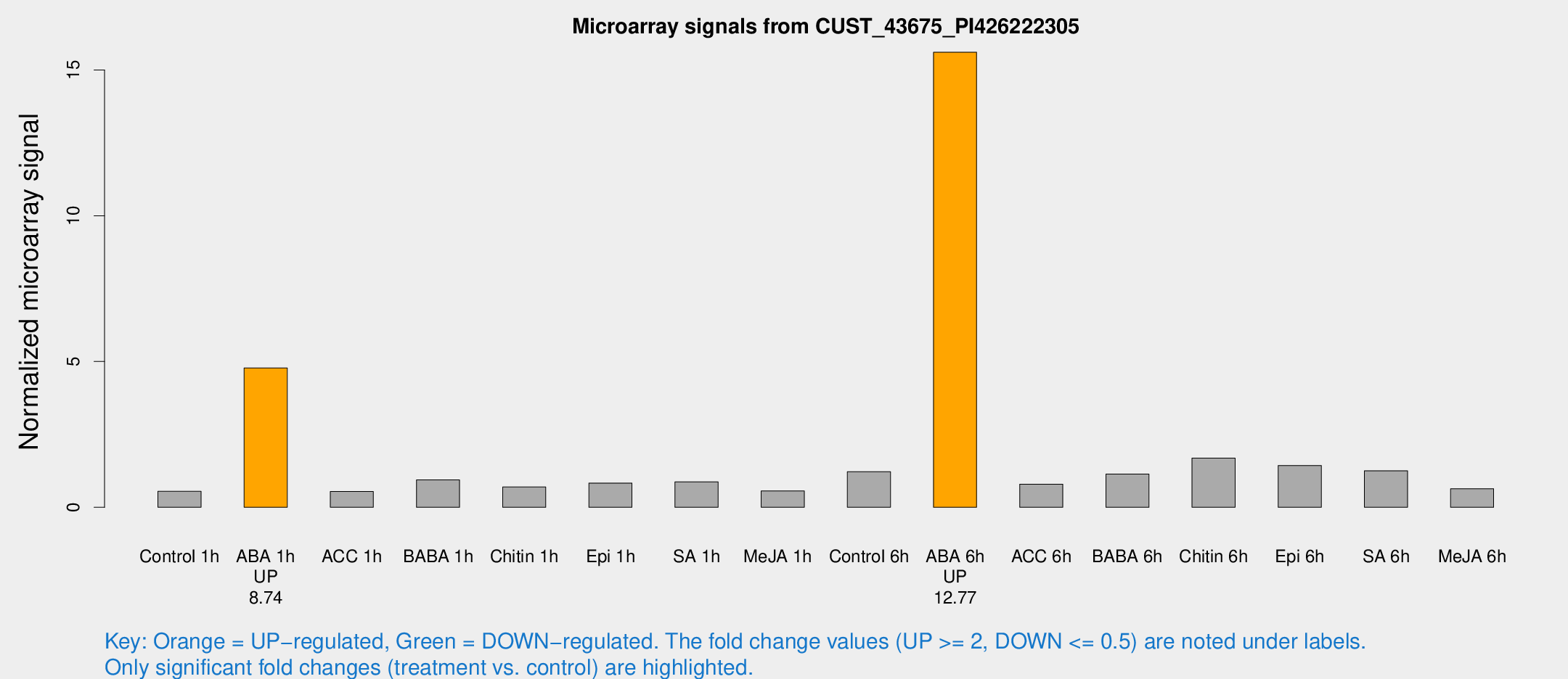
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 46.5207 | 7.88272 | 0.546667 | 0.0850755 |
| ABA 1h | 362.916 | 64.7216 | 4.77971 | 0.745251 |
| ACC 1h | 49.1454 | 11.5029 | 0.540571 | 0.110399 |
| BABA 1h | 77.4644 | 13.1377 | 0.939606 | 0.0791459 |
| Chitin 1h | 59.5587 | 21.6692 | 0.696802 | 0.260979 |
| Epi 1h | 63.6716 | 14.992 | 0.832036 | 0.227029 |
| SA 1h | 74.2417 | 5.64094 | 0.873103 | 0.0663327 |
| Me-JA 1h | 39.5951 | 8.31531 | 0.562925 | 0.0675608 |
| Control 6h | 106.411 | 24.0385 | 1.22245 | 0.214169 |
| ABA 6h | 1404.45 | 200.349 | 15.608 | 1.79978 |
| ACC 6h | 76.4845 | 8.93104 | 0.794126 | 0.0832601 |
| BABA 6h | 113.728 | 31.0824 | 1.1405 | 0.344029 |
| Chitin 6h | 149.108 | 9.59306 | 1.68729 | 0.108336 |
| Epi 6h | 134.624 | 10.768 | 1.43434 | 0.100977 |
| SA 6h | 103.535 | 10.8755 | 1.25216 | 0.103796 |
| Me-JA 6h | 70.7819 | 29.6285 | 0.637099 | 0.432723 |
Source Transcript PGSC0003DMT400004867 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G14620.1 | +3 | 0.0 | 549 | 302/554 (55%) | domains rearranged methyltransferase 2 | chr5:4715429-4718578 REVERSE LENGTH=626 |
