Probe CUST_43685_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43685_PI426222305 | JHI_St_60k_v1 | DMT400004865 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
All Microarray Probes Designed to Gene DMG400001934
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43685_PI426222305 | JHI_St_60k_v1 | DMT400004865 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43704_PI426222305 | JHI_St_60k_v1 | DMT400004864 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43690_PI426222305 | JHI_St_60k_v1 | DMT400004866 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43675_PI426222305 | JHI_St_60k_v1 | DMT400004867 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
Microarray Signals from CUST_43685_PI426222305
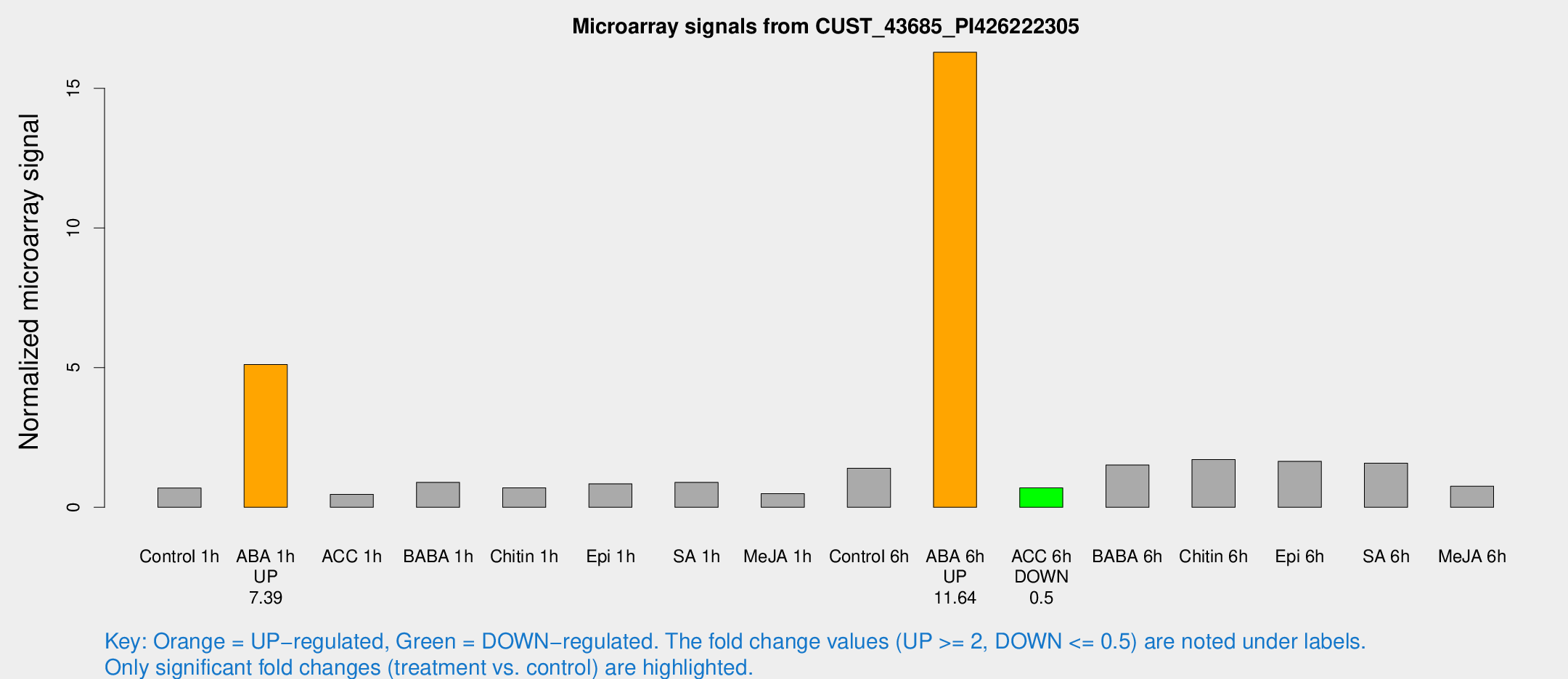
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 57.1357 | 8.67517 | 0.691482 | 0.0613825 |
| ABA 1h | 383.9 | 78.4376 | 5.11339 | 0.979119 |
| ACC 1h | 40.5871 | 8.72391 | 0.464209 | 0.0794181 |
| BABA 1h | 72.7175 | 13.8692 | 0.894558 | 0.100631 |
| Chitin 1h | 52.9186 | 11.1565 | 0.699915 | 0.0908567 |
| Epi 1h | 60.6992 | 10.493 | 0.840868 | 0.152254 |
| SA 1h | 74.3932 | 5.54484 | 0.893542 | 0.0664438 |
| Me-JA 1h | 32.871 | 5.2719 | 0.487041 | 0.0641883 |
| Control 6h | 117.065 | 21.1714 | 1.39898 | 0.1722 |
| ABA 6h | 1442.54 | 237.652 | 16.2903 | 2.24214 |
| ACC 6h | 67.4115 | 15.1102 | 0.695304 | 0.0624613 |
| BABA 6h | 143.579 | 32.6025 | 1.51242 | 0.359011 |
| Chitin 6h | 147.873 | 12.643 | 1.70875 | 0.109917 |
| Epi 6h | 151.17 | 13.2976 | 1.64754 | 0.107647 |
| SA 6h | 126.654 | 8.35967 | 1.58151 | 0.113427 |
| Me-JA 6h | 71.6656 | 26.7518 | 0.756449 | 0.278453 |
Source Transcript PGSC0003DMT400004865 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G14620.1 | +2 | 2e-175 | 530 | 266/429 (62%) | domains rearranged methyltransferase 2 | chr5:4715429-4718578 REVERSE LENGTH=626 |
