Probe CUST_43690_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43690_PI426222305 | JHI_St_60k_v1 | DMT400004866 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
All Microarray Probes Designed to Gene DMG400001934
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43685_PI426222305 | JHI_St_60k_v1 | DMT400004865 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43704_PI426222305 | JHI_St_60k_v1 | DMT400004864 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43690_PI426222305 | JHI_St_60k_v1 | DMT400004866 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
| CUST_43675_PI426222305 | JHI_St_60k_v1 | DMT400004867 | ATTTAAGTTTGGGCTAGTTAATGACTCAGTACCCATCTAAGATTTGGGCTTGAAATTATC |
Microarray Signals from CUST_43690_PI426222305
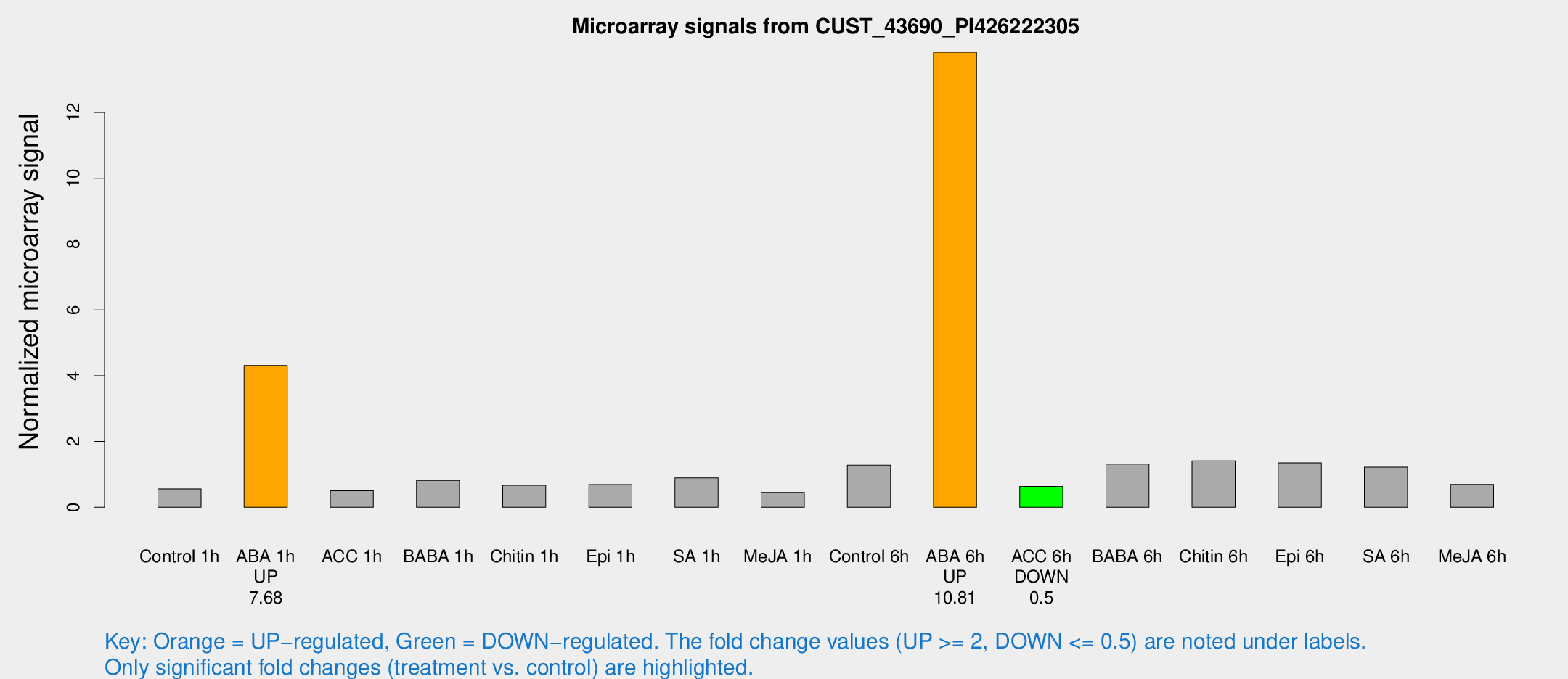
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 54.2211 | 5.04706 | 0.561222 | 0.0453149 |
| ABA 1h | 377.053 | 63.8705 | 4.31035 | 0.641391 |
| ACC 1h | 54.1322 | 14.3915 | 0.50374 | 0.122821 |
| BABA 1h | 77.8038 | 12.5009 | 0.819594 | 0.0653903 |
| Chitin 1h | 59.5684 | 11.6571 | 0.667589 | 0.0769898 |
| Epi 1h | 62.2731 | 16.2277 | 0.690429 | 0.217389 |
| SA 1h | 89.4014 | 12.4241 | 0.892237 | 0.0839518 |
| Me-JA 1h | 35.7849 | 3.6744 | 0.454866 | 0.0467186 |
| Control 6h | 127.834 | 25.0124 | 1.2791 | 0.174819 |
| ABA 6h | 1451.36 | 233.428 | 13.8314 | 1.88934 |
| ACC 6h | 73.1951 | 14.1144 | 0.638257 | 0.0873497 |
| BABA 6h | 142.38 | 14.8066 | 1.31422 | 0.122145 |
| Chitin 6h | 146.73 | 19.0141 | 1.41334 | 0.185366 |
| Epi 6h | 146.105 | 9.16146 | 1.34997 | 0.135265 |
| SA 6h | 119.184 | 20.9676 | 1.21901 | 0.113435 |
| Me-JA 6h | 75.1997 | 25.4623 | 0.693779 | 0.207955 |
Source Transcript PGSC0003DMT400004866 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G14620.1 | +2 | 2e-166 | 509 | 256/411 (62%) | domains rearranged methyltransferase 2 | chr5:4715429-4718578 REVERSE LENGTH=626 |
