Probe CUST_41182_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41182_PI426222305 | JHI_St_60k_v1 | DMT400008449 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
All Microarray Probes Designed to Gene DMG400003266
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41175_PI426222305 | JHI_St_60k_v1 | DMT400008450 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41182_PI426222305 | JHI_St_60k_v1 | DMT400008449 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41161_PI426222305 | JHI_St_60k_v1 | DMT400008448 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41174_PI426222305 | JHI_St_60k_v1 | DMT400008451 | ATTCAAAGCTAATGAACTTTGATTCTCCTTGCAGGTTAGTGTAGATTATTTAACGAAGCC |
Microarray Signals from CUST_41182_PI426222305
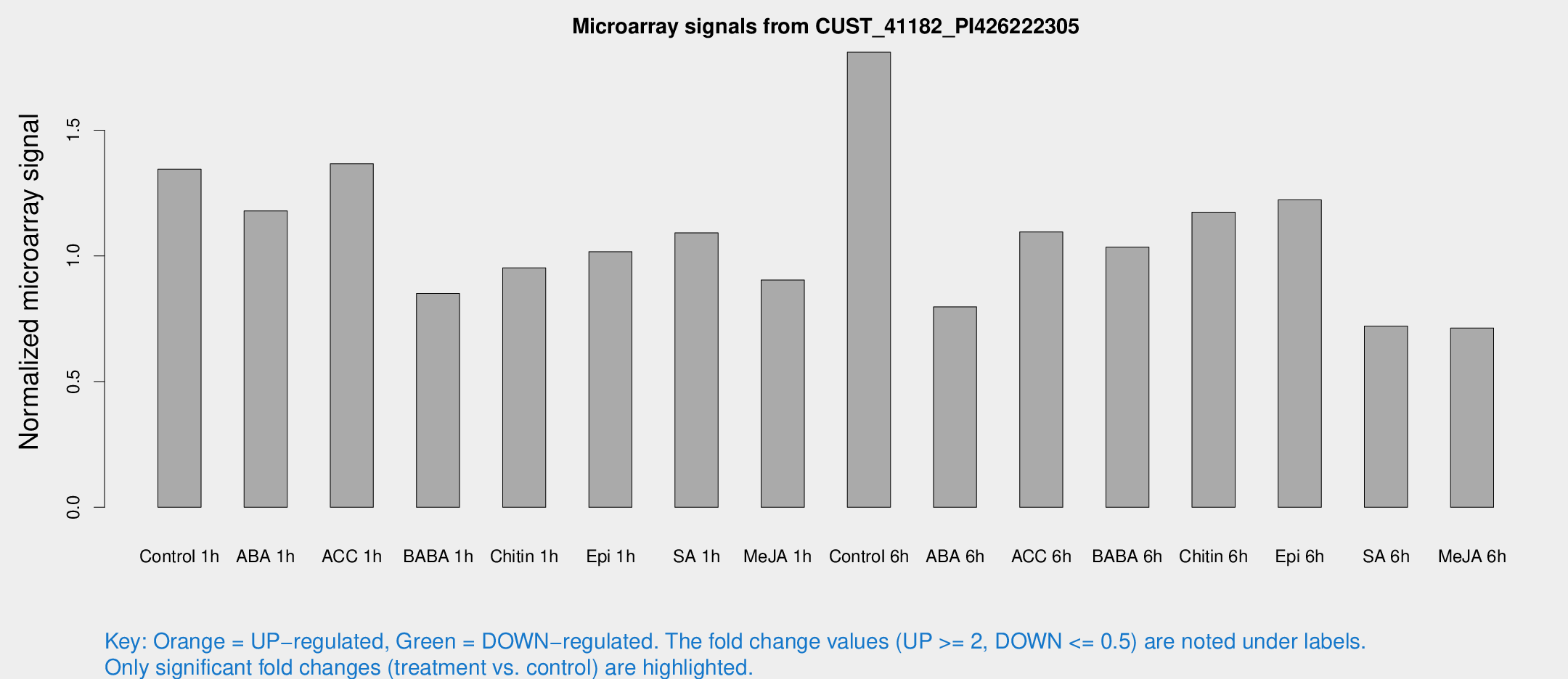
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 582.241 | 58.5545 | 1.34462 | 0.0779874 |
| ABA 1h | 456.503 | 68.1708 | 1.17907 | 0.247908 |
| ACC 1h | 608.585 | 66.8009 | 1.36634 | 0.0872616 |
| BABA 1h | 356.254 | 39.7521 | 0.850421 | 0.0497714 |
| Chitin 1h | 373.432 | 50.114 | 0.952535 | 0.148474 |
| Epi 1h | 379.206 | 26.8943 | 1.01703 | 0.0593189 |
| SA 1h | 481.329 | 28.0058 | 1.09141 | 0.0634155 |
| Me-JA 1h | 317.179 | 18.6425 | 0.903836 | 0.0610738 |
| Control 6h | 856.565 | 244.571 | 1.81022 | 0.458272 |
| ABA 6h | 366.985 | 36.8424 | 0.797494 | 0.0962384 |
| ACC 6h | 576.113 | 156.334 | 1.09542 | 0.126604 |
| BABA 6h | 513.456 | 96.7158 | 1.03501 | 0.19497 |
| Chitin 6h | 554.907 | 107.619 | 1.174 | 0.192203 |
| Epi 6h | 603.126 | 87.5529 | 1.22327 | 0.171682 |
| SA 6h | 363.311 | 124.311 | 0.721134 | 0.263499 |
| Me-JA 6h | 315.786 | 57.6327 | 0.712988 | 0.0801119 |
Source Transcript PGSC0003DMT400008449 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G29590.1 | +2 | 9e-111 | 267 | 125/147 (85%) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | chr4:14512736-14514404 REVERSE LENGTH=317 |
