Probe CUST_41174_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41174_PI426222305 | JHI_St_60k_v1 | DMT400008451 | ATTCAAAGCTAATGAACTTTGATTCTCCTTGCAGGTTAGTGTAGATTATTTAACGAAGCC |
All Microarray Probes Designed to Gene DMG400003266
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41175_PI426222305 | JHI_St_60k_v1 | DMT400008450 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41182_PI426222305 | JHI_St_60k_v1 | DMT400008449 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41161_PI426222305 | JHI_St_60k_v1 | DMT400008448 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41174_PI426222305 | JHI_St_60k_v1 | DMT400008451 | ATTCAAAGCTAATGAACTTTGATTCTCCTTGCAGGTTAGTGTAGATTATTTAACGAAGCC |
Microarray Signals from CUST_41174_PI426222305
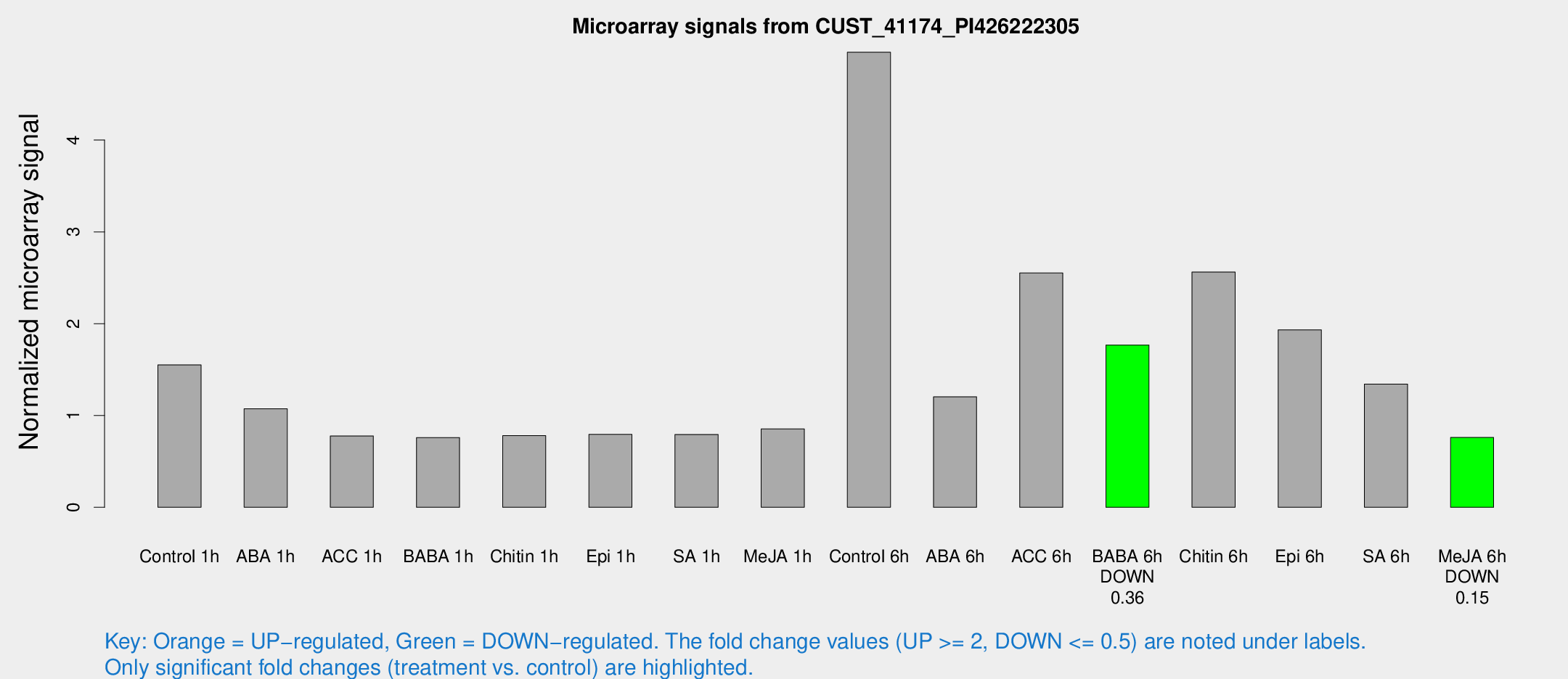
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14.01 | 5.0866 | 1.55029 | 0.699884 |
| ABA 1h | 7.59355 | 2.98013 | 1.07273 | 0.462314 |
| ACC 1h | 6.21147 | 3.64221 | 0.77633 | 0.449847 |
| BABA 1h | 5.6493 | 3.28695 | 0.759414 | 0.440886 |
| Chitin 1h | 5.27049 | 3.0759 | 0.780051 | 0.439156 |
| Epi 1h | 5.19821 | 3.02464 | 0.7941 | 0.451845 |
| SA 1h | 6.43778 | 3.1055 | 0.793698 | 0.403329 |
| Me-JA 1h | 5.39168 | 3.14015 | 0.854618 | 0.494181 |
| Control 6h | 42.1836 | 13.7527 | 4.95668 | 1.38751 |
| ABA 6h | 13.7655 | 8.10931 | 1.20352 | 0.868195 |
| ACC 6h | 23.1113 | 4.00259 | 2.55275 | 0.431973 |
| BABA 6h | 15.6037 | 3.63057 | 1.76723 | 0.435794 |
| Chitin 6h | 22.0287 | 4.9294 | 2.56221 | 0.510296 |
| Epi 6h | 17.308 | 3.80429 | 1.93256 | 0.46631 |
| SA 6h | 11.0111 | 3.41348 | 1.34153 | 0.565673 |
| Me-JA 6h | 5.84526 | 3.13414 | 0.760248 | 0.409134 |
Source Transcript PGSC0003DMT400008451 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G29590.1 | +2 | 2e-48 | 176 | 80/91 (88%) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | chr4:14512736-14514404 REVERSE LENGTH=317 |
