Probe CUST_41161_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41161_PI426222305 | JHI_St_60k_v1 | DMT400008448 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
All Microarray Probes Designed to Gene DMG400003266
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41175_PI426222305 | JHI_St_60k_v1 | DMT400008450 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41182_PI426222305 | JHI_St_60k_v1 | DMT400008449 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41161_PI426222305 | JHI_St_60k_v1 | DMT400008448 | AATTGGGGTATCGTTGTTGTCTCAGTTCATGAGTATGGCTGGCAATGTCGGTGCTAAATC |
| CUST_41174_PI426222305 | JHI_St_60k_v1 | DMT400008451 | ATTCAAAGCTAATGAACTTTGATTCTCCTTGCAGGTTAGTGTAGATTATTTAACGAAGCC |
Microarray Signals from CUST_41161_PI426222305
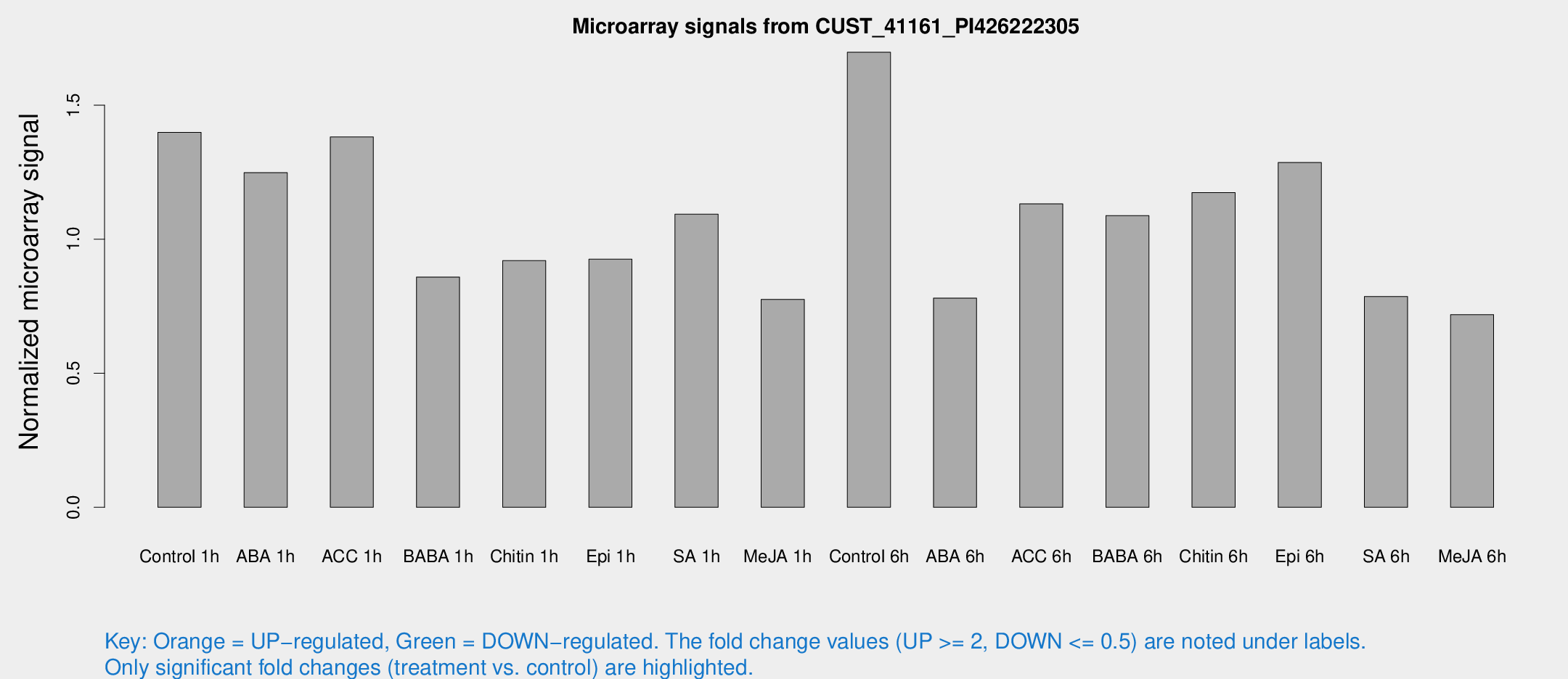
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 608.759 | 114.09 | 1.39867 | 0.163183 |
| ABA 1h | 468.869 | 44.6965 | 1.24828 | 0.198191 |
| ACC 1h | 607.833 | 77.8584 | 1.38163 | 0.0803853 |
| BABA 1h | 351.84 | 32.8368 | 0.859036 | 0.0505576 |
| Chitin 1h | 360.116 | 68.6736 | 0.920119 | 0.184706 |
| Epi 1h | 339.245 | 26.6392 | 0.92546 | 0.0543931 |
| SA 1h | 473.853 | 27.651 | 1.09335 | 0.0799263 |
| Me-JA 1h | 267.571 | 16.4894 | 0.775606 | 0.0631252 |
| Control 6h | 794.271 | 237.337 | 1.6976 | 0.453864 |
| ABA 6h | 353.176 | 38.0365 | 0.780467 | 0.0884801 |
| ACC 6h | 597.885 | 188.475 | 1.13203 | 0.177952 |
| BABA 6h | 533.858 | 111.775 | 1.08814 | 0.208581 |
| Chitin 6h | 553.319 | 129.101 | 1.17397 | 0.22829 |
| Epi 6h | 614.608 | 51.7334 | 1.28609 | 0.145081 |
| SA 6h | 382.092 | 125.701 | 0.786292 | 0.260542 |
| Me-JA 6h | 313.704 | 60.1262 | 0.718587 | 0.0879025 |
Source Transcript PGSC0003DMT400008448 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G29590.1 | +3 | 2e-78 | 244 | 121/179 (68%) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | chr4:14512736-14514404 REVERSE LENGTH=317 |
