Probe CUST_36319_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36319_PI426222305 | JHI_St_60k_v1 | DMT400039810 | ACAGAAAAGTATCGATCAATAGTTTTTAAGGAACCCCGATTTGTTGAGTATTTCCGCCTG |
All Microarray Probes Designed to Gene DMG400015385
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36315_PI426222305 | JHI_St_60k_v1 | DMT400039808 | TCAATTCTCCTCTTGGATGGGCGGTGATCGTGATGGTCCAAGATTGTTATGAACTTTCTG |
| CUST_36319_PI426222305 | JHI_St_60k_v1 | DMT400039810 | ACAGAAAAGTATCGATCAATAGTTTTTAAGGAACCCCGATTTGTTGAGTATTTCCGCCTG |
| CUST_36250_PI426222305 | JHI_St_60k_v1 | DMT400039807 | CTGCTACCAAATACAGGTAATCCAAGAGTGACTCCTGAGGTCACAAGAGATGTTTGCTTA |
| CUST_36249_PI426222305 | JHI_St_60k_v1 | DMT400039809 | CAAGAACTTGATGAGGCTTTACAGAGGGAGGTCCAAGATTGTTATGAACTTTCTGCCGAG |
Microarray Signals from CUST_36319_PI426222305
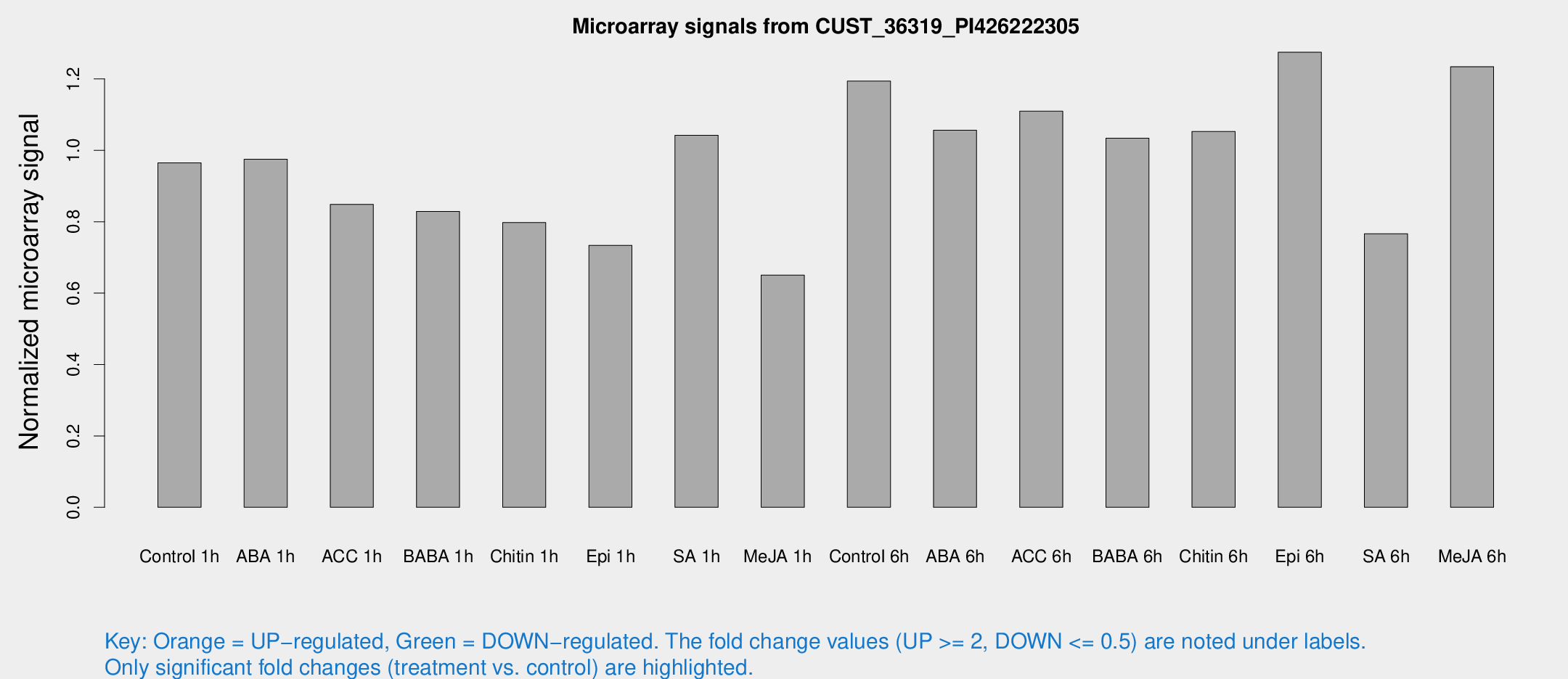
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2064.3 | 220.586 | 0.964957 | 0.0557284 |
| ABA 1h | 1836.96 | 153.101 | 0.974907 | 0.0563057 |
| ACC 1h | 1922.59 | 360.736 | 0.848709 | 0.1134 |
| BABA 1h | 1806.2 | 425.862 | 0.828959 | 0.140376 |
| Chitin 1h | 1540.5 | 193.559 | 0.797628 | 0.0634232 |
| Epi 1h | 1364.24 | 161.093 | 0.73398 | 0.0861006 |
| SA 1h | 2289.66 | 238.029 | 1.04244 | 0.0782109 |
| Me-JA 1h | 1125.42 | 65.1197 | 0.650681 | 0.037605 |
| Control 6h | 2683.98 | 601.52 | 1.194 | 0.203486 |
| ABA 6h | 2432.27 | 348.444 | 1.05634 | 0.124244 |
| ACC 6h | 2730.31 | 327.997 | 1.10961 | 0.064079 |
| BABA 6h | 2482.31 | 286.528 | 1.03404 | 0.0979444 |
| Chitin 6h | 2372.37 | 137.085 | 1.05263 | 0.0607908 |
| Epi 6h | 3084.09 | 365.294 | 1.2749 | 0.130757 |
| SA 6h | 2072.53 | 817.979 | 0.766491 | 0.403945 |
| Me-JA 6h | 2766.5 | 622.054 | 1.23431 | 0.225452 |
Source Transcript PGSC0003DMT400039810 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G53310.1 | +1 | 0.0 | 968 | 499/560 (89%) | phosphoenolpyruvate carboxylase 1 | chr1:19884261-19888070 REVERSE LENGTH=967 |
