Probe CUST_36315_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36315_PI426222305 | JHI_St_60k_v1 | DMT400039808 | TCAATTCTCCTCTTGGATGGGCGGTGATCGTGATGGTCCAAGATTGTTATGAACTTTCTG |
All Microarray Probes Designed to Gene DMG400015385
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36315_PI426222305 | JHI_St_60k_v1 | DMT400039808 | TCAATTCTCCTCTTGGATGGGCGGTGATCGTGATGGTCCAAGATTGTTATGAACTTTCTG |
| CUST_36319_PI426222305 | JHI_St_60k_v1 | DMT400039810 | ACAGAAAAGTATCGATCAATAGTTTTTAAGGAACCCCGATTTGTTGAGTATTTCCGCCTG |
| CUST_36250_PI426222305 | JHI_St_60k_v1 | DMT400039807 | CTGCTACCAAATACAGGTAATCCAAGAGTGACTCCTGAGGTCACAAGAGATGTTTGCTTA |
| CUST_36249_PI426222305 | JHI_St_60k_v1 | DMT400039809 | CAAGAACTTGATGAGGCTTTACAGAGGGAGGTCCAAGATTGTTATGAACTTTCTGCCGAG |
Microarray Signals from CUST_36315_PI426222305
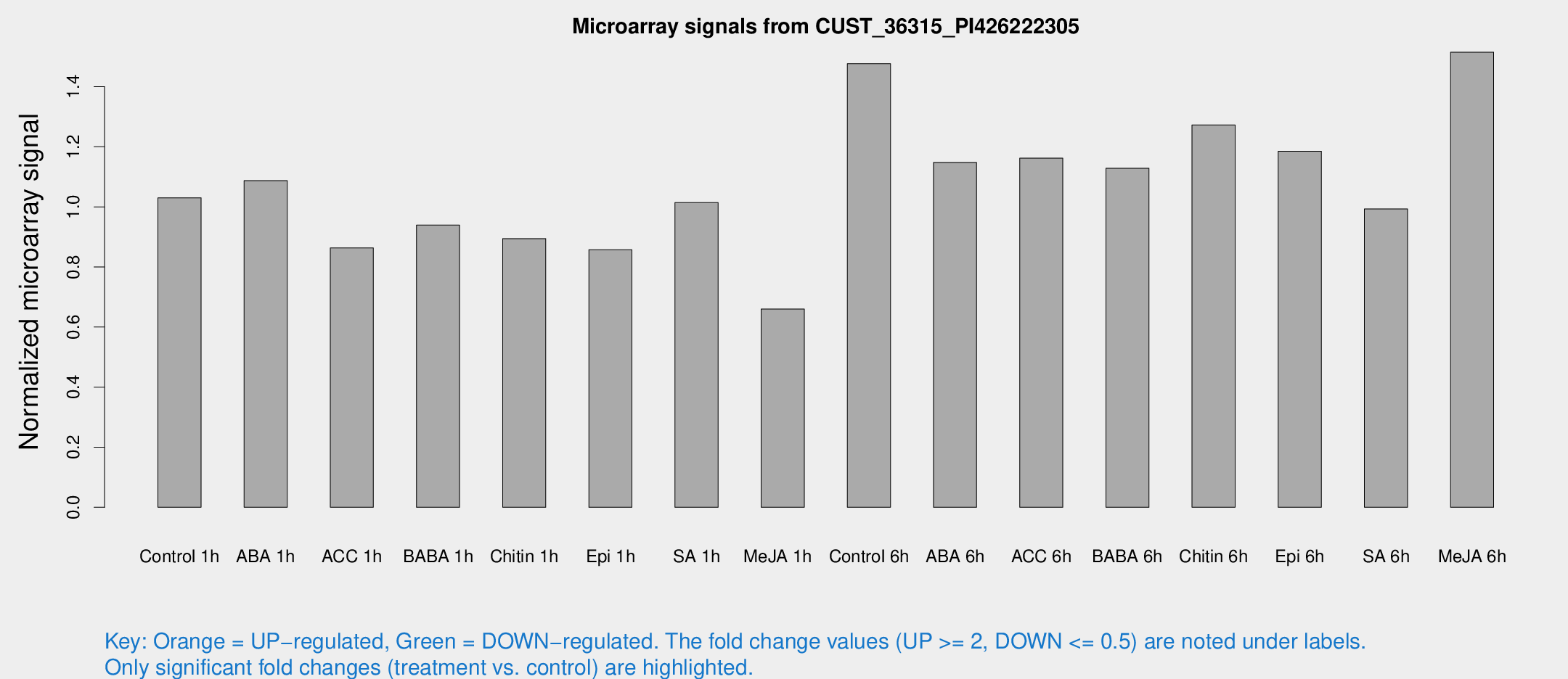
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 318.185 | 45.0815 | 1.03022 | 0.0753082 |
| ABA 1h | 292.418 | 17.6058 | 1.08724 | 0.0759216 |
| ACC 1h | 280.402 | 53.2081 | 0.86319 | 0.120012 |
| BABA 1h | 287.412 | 60.5456 | 0.939249 | 0.124262 |
| Chitin 1h | 245.08 | 18.9192 | 0.894415 | 0.0534209 |
| Epi 1h | 232.238 | 38.783 | 0.857377 | 0.150758 |
| SA 1h | 318.971 | 32.349 | 1.014 | 0.0597314 |
| Me-JA 1h | 164.233 | 12.8638 | 0.659732 | 0.0409785 |
| Control 6h | 493.224 | 136.936 | 1.47663 | 0.369136 |
| ABA 6h | 398.431 | 98.6264 | 1.14795 | 0.281515 |
| ACC 6h | 415.63 | 68.8562 | 1.16193 | 0.0939711 |
| BABA 6h | 399.067 | 75.9866 | 1.12851 | 0.205574 |
| Chitin 6h | 421.863 | 65.6812 | 1.2724 | 0.229756 |
| Epi 6h | 430.054 | 108.78 | 1.18535 | 0.332026 |
| SA 6h | 314.582 | 71.7973 | 0.993039 | 0.294621 |
| Me-JA 6h | 499.885 | 129.706 | 1.51465 | 0.389661 |
Source Transcript PGSC0003DMT400039808 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G14940.1 | +1 | 0.0 | 1305 | 660/752 (88%) | phosphoenolpyruvate carboxylase 3 | chr3:5025584-5029476 FORWARD LENGTH=968 |
