Probe CUST_36249_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36249_PI426222305 | JHI_St_60k_v1 | DMT400039809 | CAAGAACTTGATGAGGCTTTACAGAGGGAGGTCCAAGATTGTTATGAACTTTCTGCCGAG |
All Microarray Probes Designed to Gene DMG400015385
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36315_PI426222305 | JHI_St_60k_v1 | DMT400039808 | TCAATTCTCCTCTTGGATGGGCGGTGATCGTGATGGTCCAAGATTGTTATGAACTTTCTG |
| CUST_36319_PI426222305 | JHI_St_60k_v1 | DMT400039810 | ACAGAAAAGTATCGATCAATAGTTTTTAAGGAACCCCGATTTGTTGAGTATTTCCGCCTG |
| CUST_36250_PI426222305 | JHI_St_60k_v1 | DMT400039807 | CTGCTACCAAATACAGGTAATCCAAGAGTGACTCCTGAGGTCACAAGAGATGTTTGCTTA |
| CUST_36249_PI426222305 | JHI_St_60k_v1 | DMT400039809 | CAAGAACTTGATGAGGCTTTACAGAGGGAGGTCCAAGATTGTTATGAACTTTCTGCCGAG |
Microarray Signals from CUST_36249_PI426222305
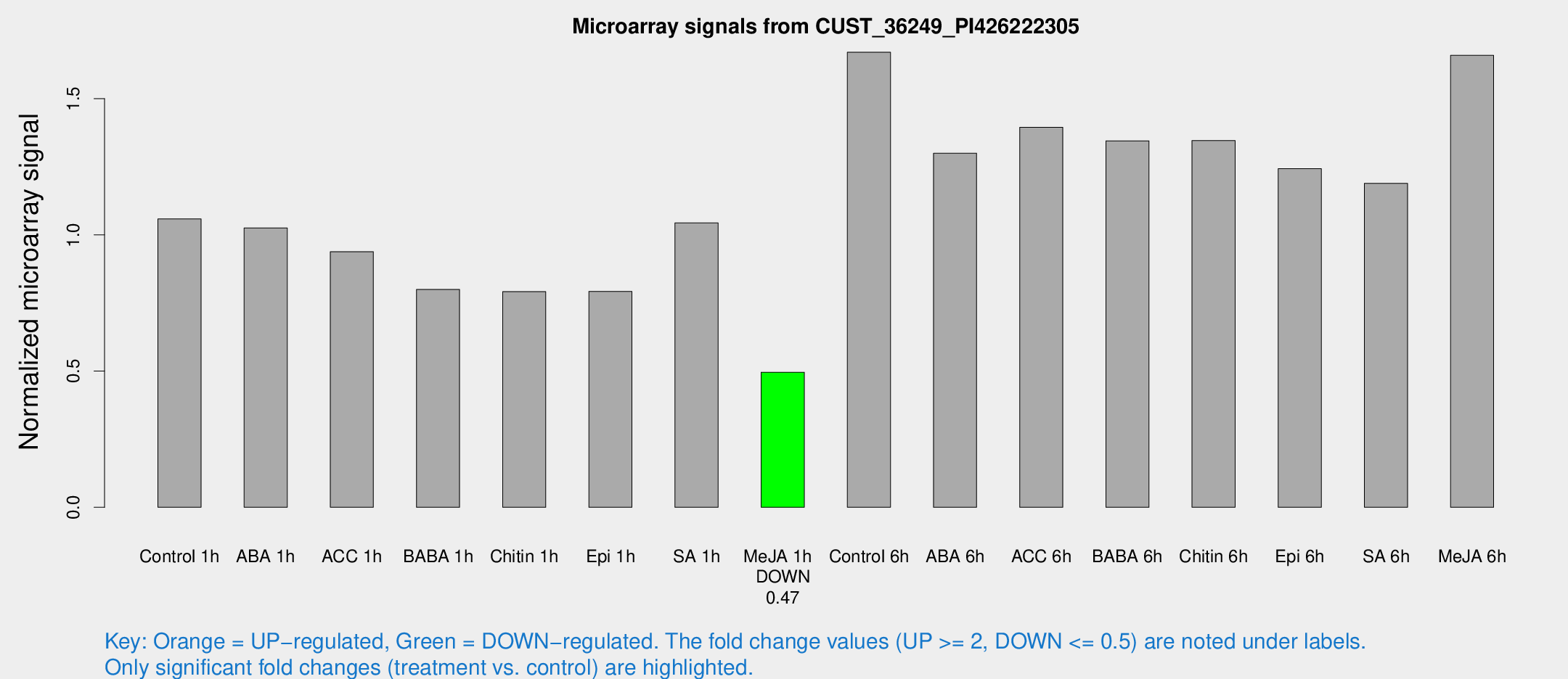
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 66.7024 | 10.466 | 1.0588 | 0.0939066 |
| ABA 1h | 55.8598 | 4.39226 | 1.0256 | 0.102221 |
| ACC 1h | 61.0812 | 10.4461 | 0.938344 | 0.111437 |
| BABA 1h | 50.481 | 11.5148 | 0.799863 | 0.128816 |
| Chitin 1h | 44.5745 | 6.00505 | 0.791866 | 0.0842197 |
| Epi 1h | 42.5371 | 3.97807 | 0.792557 | 0.0750338 |
| SA 1h | 68.1851 | 12.8615 | 1.04411 | 0.12877 |
| Me-JA 1h | 25.0985 | 3.49559 | 0.495891 | 0.069274 |
| Control 6h | 114.936 | 33.0921 | 1.67075 | 0.465371 |
| ABA 6h | 96.5333 | 28.7418 | 1.29988 | 0.489157 |
| ACC 6h | 102.85 | 21.8541 | 1.39517 | 0.114066 |
| BABA 6h | 98.433 | 23.0784 | 1.34539 | 0.31221 |
| Chitin 6h | 92.1213 | 17.6649 | 1.34629 | 0.306713 |
| Epi 6h | 95.5626 | 32.2983 | 1.24328 | 0.42163 |
| SA 6h | 81.7535 | 24.5939 | 1.18911 | 0.598753 |
| Me-JA 6h | 122.666 | 42.7057 | 1.65911 | 0.748871 |
Source Transcript PGSC0003DMT400039809 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G53310.1 | +2 | 0.0 | 1676 | 850/967 (88%) | phosphoenolpyruvate carboxylase 1 | chr1:19884261-19888070 REVERSE LENGTH=967 |
