Probe CUST_36020_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36020_PI426222305 | JHI_St_60k_v1 | DMT400080933 | GGATTTGACACACACCTGACGATATTTTTGAAGAGTACGAGCAACAAAACATTTAAATCA |
All Microarray Probes Designed to Gene DMG400031522
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35962_PI426222305 | JHI_St_60k_v1 | DMT400080934 | ATGCAGAGTGGAGAAGCAAAAAATGAGGCAGCACCAATGGCTATAGATAGTAAGGCTTAA |
| CUST_36003_PI426222305 | JHI_St_60k_v1 | DMT400080935 | CCAAGTACATGACACAAATTATGGAATTAATGTCTCTTCCATAGTGTGCAGTCAAAAAGA |
| CUST_35973_PI426222305 | JHI_St_60k_v1 | DMT400080932 | TGGTTCCGCTGCTGTTGGGAAGAGTAAGACGTCTTTTATTAATCATCAAAAAACTAAACA |
| CUST_36020_PI426222305 | JHI_St_60k_v1 | DMT400080933 | GGATTTGACACACACCTGACGATATTTTTGAAGAGTACGAGCAACAAAACATTTAAATCA |
Microarray Signals from CUST_36020_PI426222305
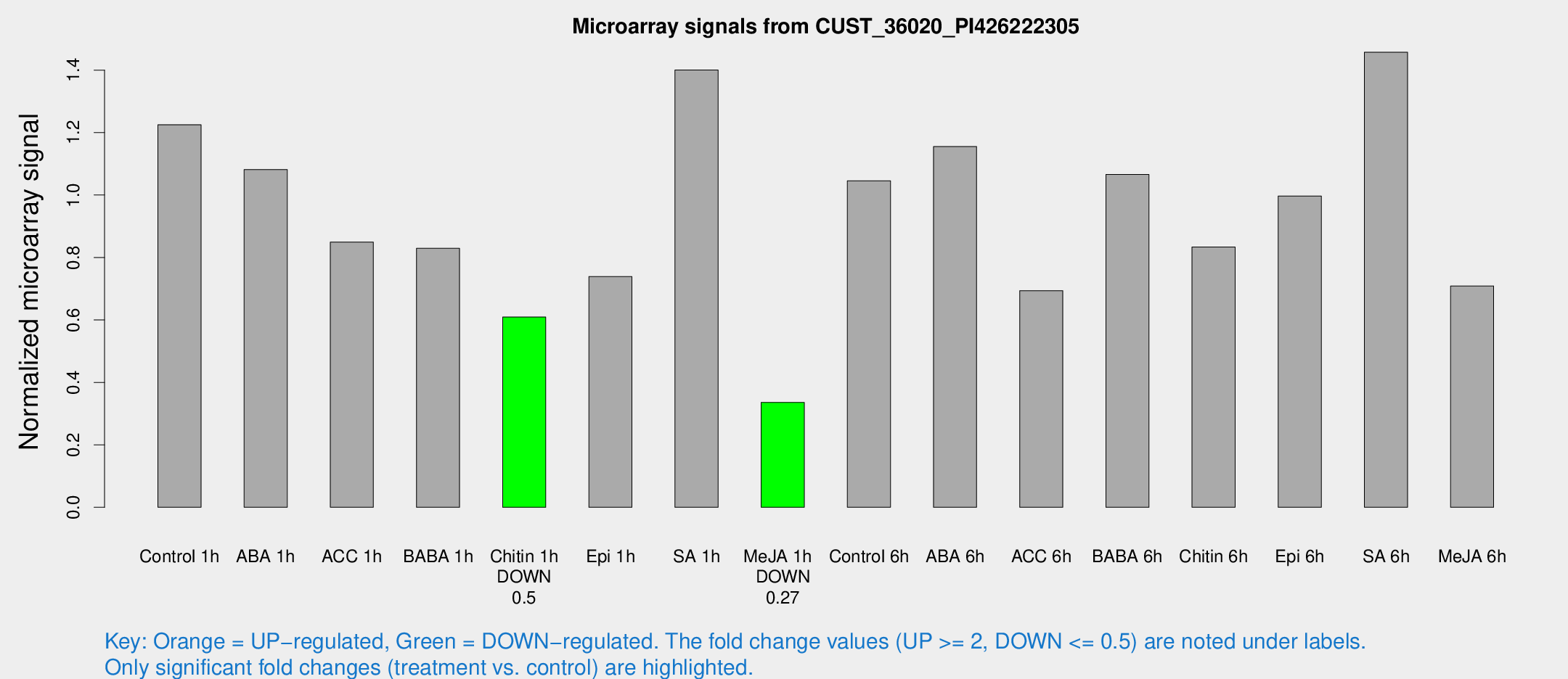
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 99.4524 | 15.5516 | 1.22505 | 0.15992 |
| ABA 1h | 76.1618 | 5.57081 | 1.08177 | 0.0795478 |
| ACC 1h | 80.9676 | 26.0939 | 0.84942 | 0.335923 |
| BABA 1h | 67.3865 | 14.9747 | 0.829396 | 0.131423 |
| Chitin 1h | 45.2076 | 9.4915 | 0.609351 | 0.0859405 |
| Epi 1h | 52.256 | 9.49965 | 0.738891 | 0.109388 |
| SA 1h | 119.702 | 27.0303 | 1.4003 | 0.225515 |
| Me-JA 1h | 22.1626 | 3.78935 | 0.335668 | 0.0589227 |
| Control 6h | 85.4743 | 13.5102 | 1.04576 | 0.0833633 |
| ABA 6h | 100.203 | 17.7963 | 1.15529 | 0.136339 |
| ACC 6h | 63.7518 | 6.50322 | 0.693841 | 0.104546 |
| BABA 6h | 97.397 | 17.1476 | 1.06593 | 0.167593 |
| Chitin 6h | 73.1821 | 13.4069 | 0.833629 | 0.155266 |
| Epi 6h | 91.3848 | 14.3605 | 0.996845 | 0.0853756 |
| SA 6h | 114.699 | 7.6184 | 1.45731 | 0.0969612 |
| Me-JA 6h | 63.7031 | 24.209 | 0.708633 | 0.212207 |
Source Transcript PGSC0003DMT400080933 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G25230.2 | +1 | 0.0 | 649 | 361/468 (77%) | rotamase FKBP 1 | chr3:9188257-9191175 FORWARD LENGTH=562 |
