Probe CUST_36003_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36003_PI426222305 | JHI_St_60k_v1 | DMT400080935 | CCAAGTACATGACACAAATTATGGAATTAATGTCTCTTCCATAGTGTGCAGTCAAAAAGA |
All Microarray Probes Designed to Gene DMG400031522
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35962_PI426222305 | JHI_St_60k_v1 | DMT400080934 | ATGCAGAGTGGAGAAGCAAAAAATGAGGCAGCACCAATGGCTATAGATAGTAAGGCTTAA |
| CUST_36003_PI426222305 | JHI_St_60k_v1 | DMT400080935 | CCAAGTACATGACACAAATTATGGAATTAATGTCTCTTCCATAGTGTGCAGTCAAAAAGA |
| CUST_35973_PI426222305 | JHI_St_60k_v1 | DMT400080932 | TGGTTCCGCTGCTGTTGGGAAGAGTAAGACGTCTTTTATTAATCATCAAAAAACTAAACA |
| CUST_36020_PI426222305 | JHI_St_60k_v1 | DMT400080933 | GGATTTGACACACACCTGACGATATTTTTGAAGAGTACGAGCAACAAAACATTTAAATCA |
Microarray Signals from CUST_36003_PI426222305
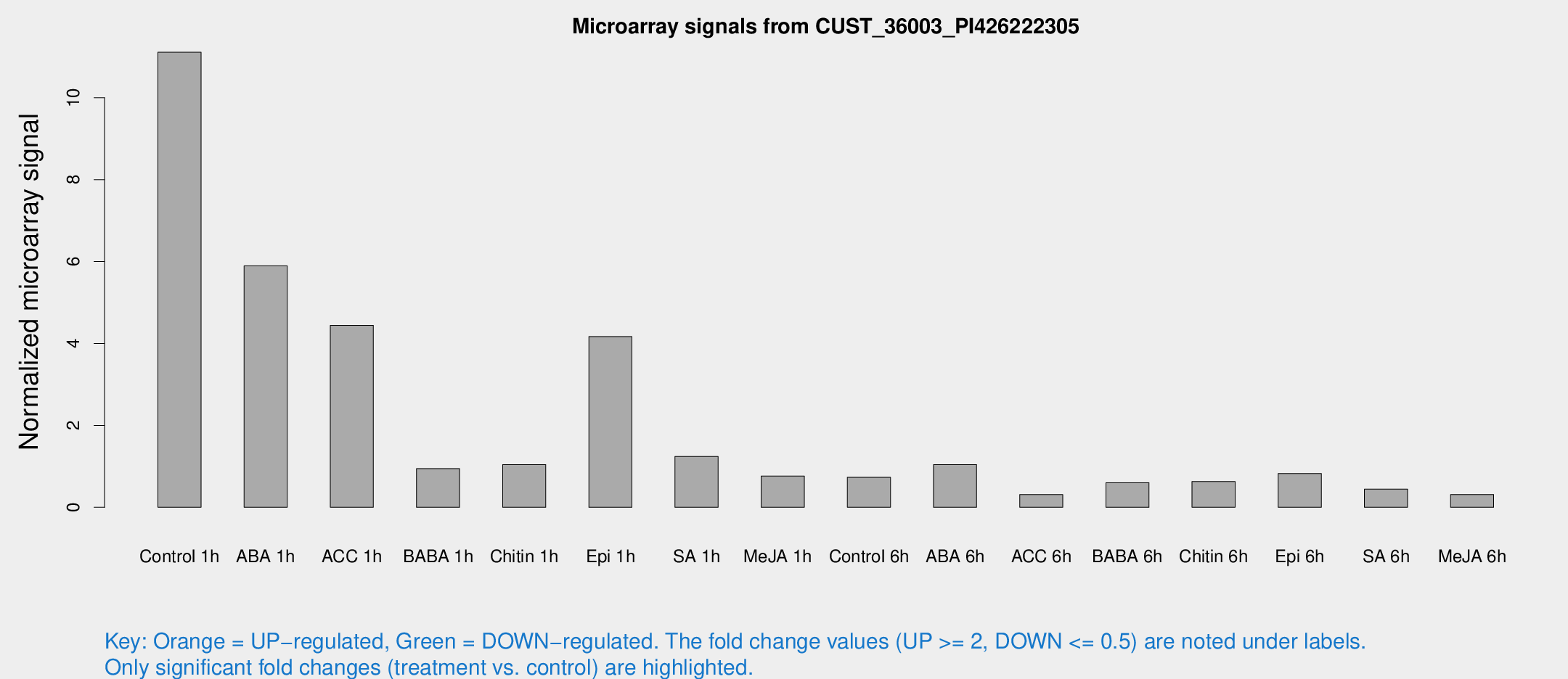
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 284.531 | 69.1201 | 11.1124 | 2.60237 |
| ABA 1h | 340.558 | 286.697 | 5.89615 | 18.838 |
| ACC 1h | 195.477 | 126.12 | 4.44169 | 5.60662 |
| BABA 1h | 31.2669 | 18.8757 | 0.944931 | 0.641303 |
| Chitin 1h | 27.2363 | 12.3331 | 1.04393 | 0.399182 |
| Epi 1h | 99.7261 | 36.5022 | 4.16933 | 1.85387 |
| SA 1h | 32.3302 | 6.63769 | 1.24192 | 0.236823 |
| Me-JA 1h | 17.6618 | 5.8468 | 0.76099 | 0.400949 |
| Control 6h | 21.2983 | 7.30777 | 0.734224 | 0.311369 |
| ABA 6h | 30.1107 | 10.2893 | 1.04218 | 0.445965 |
| ACC 6h | 9.1603 | 4.6491 | 0.311202 | 0.163946 |
| BABA 6h | 18.4239 | 6.5238 | 0.600925 | 0.251286 |
| Chitin 6h | 17.4048 | 5.02362 | 0.629817 | 0.190236 |
| Epi 6h | 23.4589 | 4.80197 | 0.824566 | 0.260291 |
| SA 6h | 11.2584 | 4.20274 | 0.445326 | 0.190744 |
| Me-JA 6h | 7.48166 | 3.98208 | 0.311831 | 0.166299 |
Source Transcript PGSC0003DMT400080935 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G48570.1 | +1 | 0.0 | 703 | 389/519 (75%) | FKBP-type peptidyl-prolyl cis-trans isomerase family protein | chr5:19690746-19693656 REVERSE LENGTH=578 |
