Probe CUST_35962_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35962_PI426222305 | JHI_St_60k_v1 | DMT400080934 | ATGCAGAGTGGAGAAGCAAAAAATGAGGCAGCACCAATGGCTATAGATAGTAAGGCTTAA |
All Microarray Probes Designed to Gene DMG400031522
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_35962_PI426222305 | JHI_St_60k_v1 | DMT400080934 | ATGCAGAGTGGAGAAGCAAAAAATGAGGCAGCACCAATGGCTATAGATAGTAAGGCTTAA |
| CUST_36003_PI426222305 | JHI_St_60k_v1 | DMT400080935 | CCAAGTACATGACACAAATTATGGAATTAATGTCTCTTCCATAGTGTGCAGTCAAAAAGA |
| CUST_35973_PI426222305 | JHI_St_60k_v1 | DMT400080932 | TGGTTCCGCTGCTGTTGGGAAGAGTAAGACGTCTTTTATTAATCATCAAAAAACTAAACA |
| CUST_36020_PI426222305 | JHI_St_60k_v1 | DMT400080933 | GGATTTGACACACACCTGACGATATTTTTGAAGAGTACGAGCAACAAAACATTTAAATCA |
Microarray Signals from CUST_35962_PI426222305
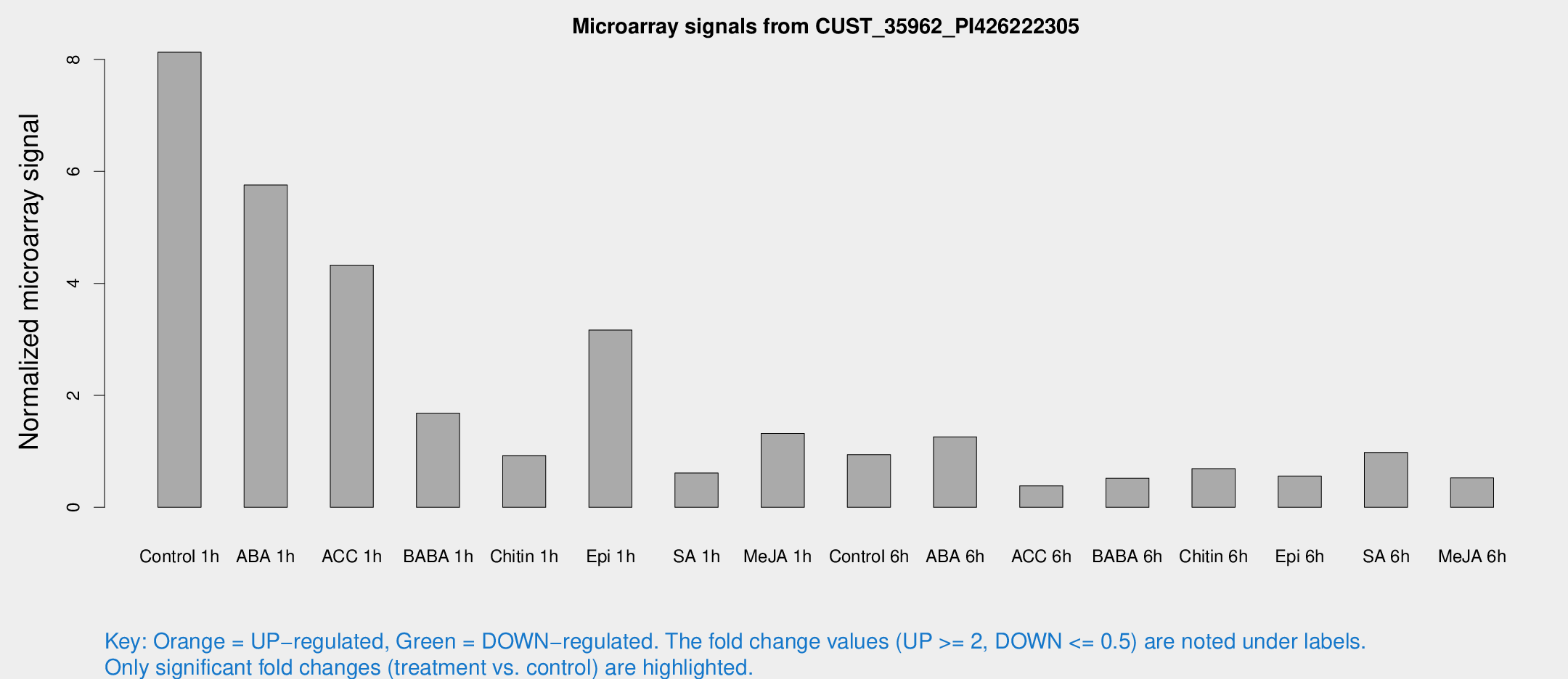
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 254.599 | 68.4146 | 8.12904 | 2.10331 |
| ABA 1h | 278.665 | 193.573 | 5.75745 | 10.039 |
| ACC 1h | 180.337 | 97.4353 | 4.32695 | 2.85851 |
| BABA 1h | 50.4493 | 11.7893 | 1.6827 | 0.52493 |
| Chitin 1h | 26.5961 | 7.86287 | 0.924526 | 0.21275 |
| Epi 1h | 91.376 | 30.0301 | 3.16654 | 1.44654 |
| SA 1h | 20.3204 | 6.09161 | 0.612911 | 0.188853 |
| Me-JA 1h | 35.6086 | 12.8859 | 1.31854 | 0.651472 |
| Control 6h | 43.8552 | 22.2672 | 0.940291 | 1.00611 |
| ABA 6h | 40.0988 | 6.10274 | 1.25932 | 0.133974 |
| ACC 6h | 13.106 | 4.02933 | 0.382741 | 0.11672 |
| BABA 6h | 17.488 | 3.85619 | 0.520504 | 0.126509 |
| Chitin 6h | 21.8318 | 3.82574 | 0.690652 | 0.125026 |
| Epi 6h | 21.5455 | 6.97913 | 0.556889 | 0.341005 |
| SA 6h | 34.7408 | 14.7004 | 0.980213 | 0.726642 |
| Me-JA 6h | 16.4236 | 4.56943 | 0.525255 | 0.128118 |
Source Transcript PGSC0003DMT400080934 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G25230.2 | +1 | 0.0 | 696 | 382/500 (76%) | rotamase FKBP 1 | chr3:9188257-9191175 FORWARD LENGTH=562 |
