Probe CUST_30350_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30350_PI426222305 | JHI_St_60k_v1 | DMT400069917 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
All Microarray Probes Designed to Gene DMG400027187
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30435_PI426222305 | JHI_St_60k_v1 | DMT400069918 | GGAGGGAAGTGTCCACTTTGACTCTTGAATCTAAACTTAGTGCAACATTTAACTAAAGAG |
| CUST_30429_PI426222305 | JHI_St_60k_v1 | DMT400069915 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
| CUST_30415_PI426222305 | JHI_St_60k_v1 | DMT400069916 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
| CUST_30350_PI426222305 | JHI_St_60k_v1 | DMT400069917 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
| CUST_30375_PI426222305 | JHI_St_60k_v1 | DMT400069914 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
Microarray Signals from CUST_30350_PI426222305
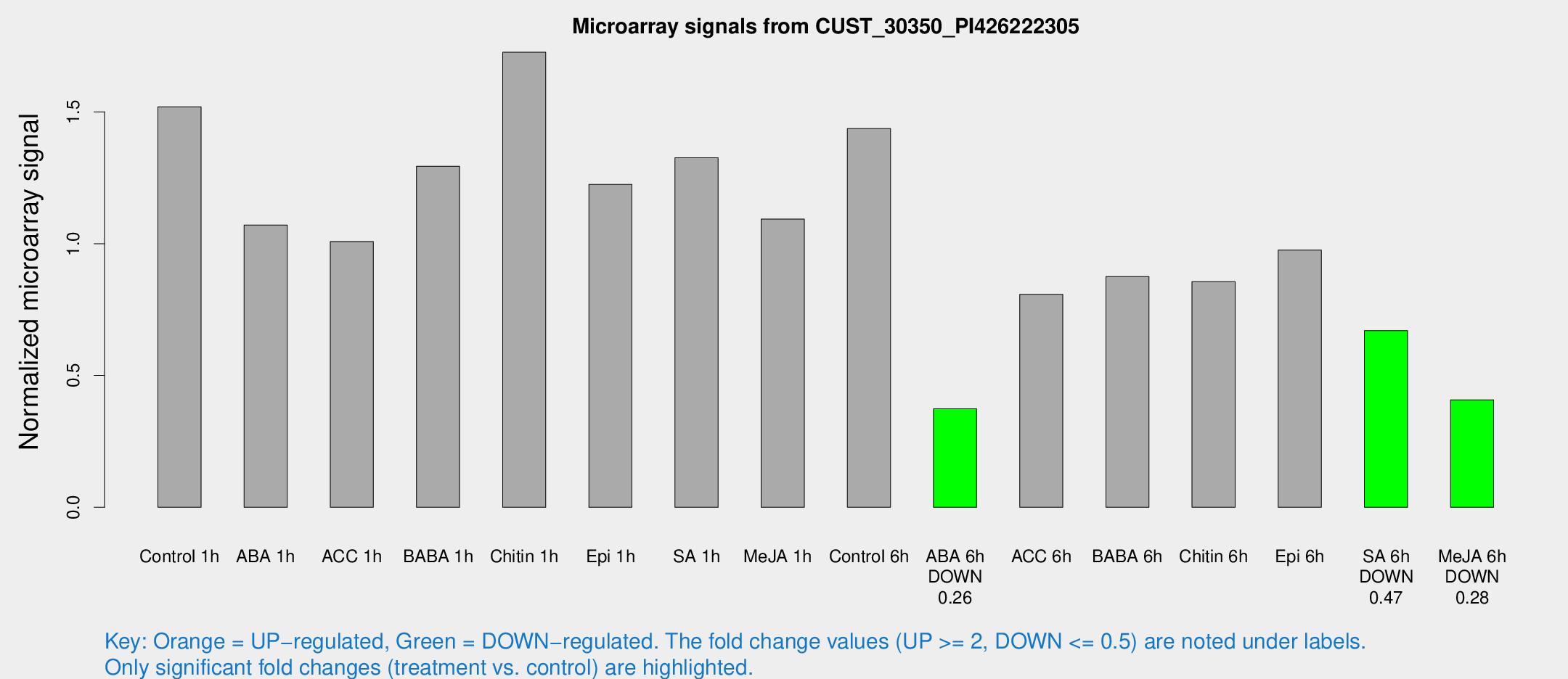
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2080.75 | 175.069 | 1.51915 | 0.0877345 |
| ABA 1h | 1323.99 | 208.242 | 1.07078 | 0.177899 |
| ACC 1h | 1446.33 | 221.216 | 1.00814 | 0.0905171 |
| BABA 1h | 1825.51 | 437.824 | 1.29349 | 0.240091 |
| Chitin 1h | 2115.49 | 122.438 | 1.72609 | 0.136136 |
| Epi 1h | 1485.5 | 253.187 | 1.22492 | 0.186557 |
| SA 1h | 1889.07 | 256.544 | 1.32589 | 0.107319 |
| Me-JA 1h | 1230.62 | 149.532 | 1.09301 | 0.0631632 |
| Control 6h | 2054.43 | 419.129 | 1.4365 | 0.207945 |
| ABA 6h | 550.143 | 75.7424 | 0.373492 | 0.0299224 |
| ACC 6h | 1277.75 | 143.601 | 0.807668 | 0.0896735 |
| BABA 6h | 1332.81 | 77.0513 | 0.875119 | 0.050574 |
| Chitin 6h | 1258.56 | 149.79 | 0.855818 | 0.0894318 |
| Epi 6h | 1536.81 | 252.817 | 0.97545 | 0.135838 |
| SA 6h | 958.559 | 217.255 | 0.669939 | 0.0969048 |
| Me-JA 6h | 593.039 | 145.102 | 0.407627 | 0.0817603 |
Source Transcript PGSC0003DMT400069917 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G39890.1 | +2 | 0.0 | 541 | 288/420 (69%) | proline transporter 1 | chr2:16656022-16658202 FORWARD LENGTH=442 |
