Probe CUST_30435_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30435_PI426222305 | JHI_St_60k_v1 | DMT400069918 | GGAGGGAAGTGTCCACTTTGACTCTTGAATCTAAACTTAGTGCAACATTTAACTAAAGAG |
All Microarray Probes Designed to Gene DMG400027187
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30435_PI426222305 | JHI_St_60k_v1 | DMT400069918 | GGAGGGAAGTGTCCACTTTGACTCTTGAATCTAAACTTAGTGCAACATTTAACTAAAGAG |
| CUST_30429_PI426222305 | JHI_St_60k_v1 | DMT400069915 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
| CUST_30415_PI426222305 | JHI_St_60k_v1 | DMT400069916 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
| CUST_30350_PI426222305 | JHI_St_60k_v1 | DMT400069917 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
| CUST_30375_PI426222305 | JHI_St_60k_v1 | DMT400069914 | CTGCATTGAGGTTCATCACAGTAGATTCCAAAACATACCATGTATTTGCTGACTTATAAT |
Microarray Signals from CUST_30435_PI426222305
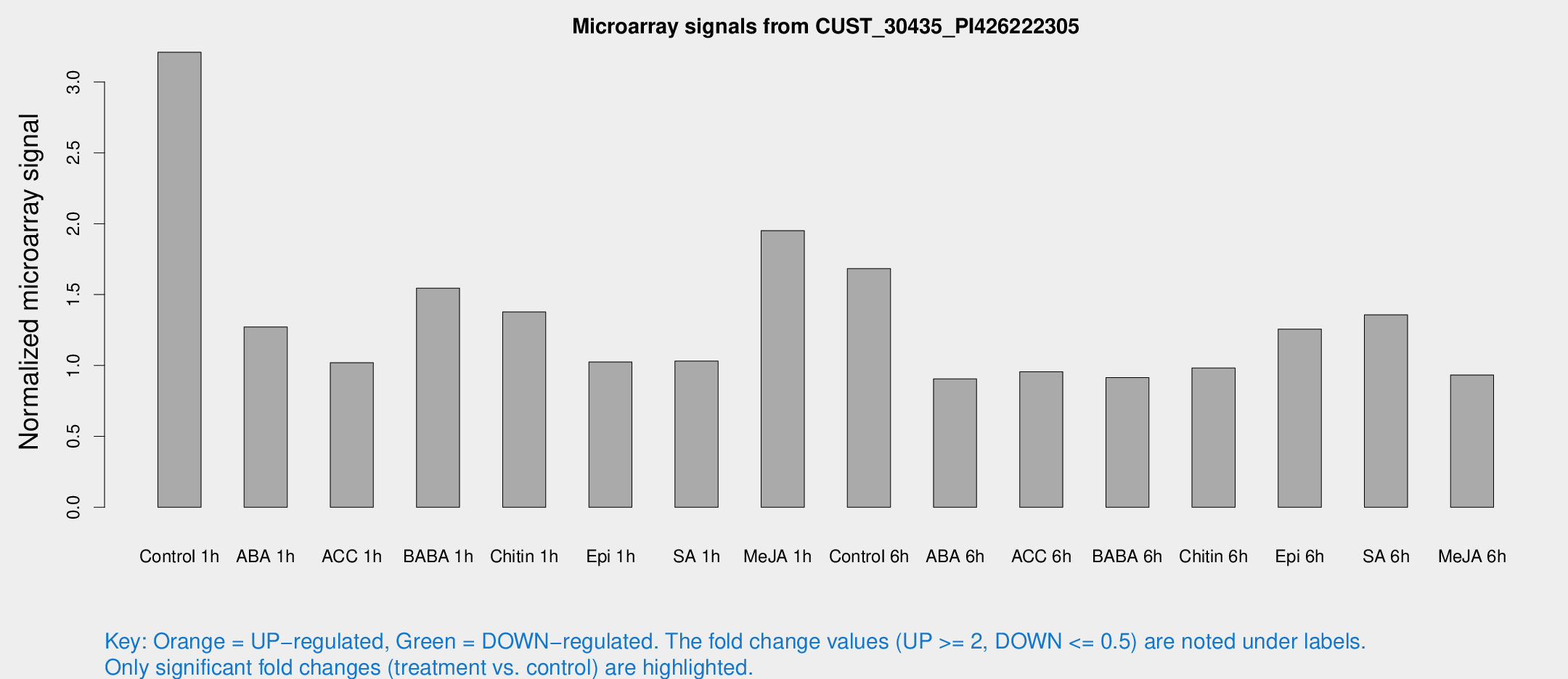
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 72.0701 | 65.7692 | 3.20927 | 14.2427 |
| ABA 1h | 7.42261 | 3.15336 | 1.27224 | 0.614392 |
| ACC 1h | 6.61255 | 3.88325 | 1.01937 | 0.590337 |
| BABA 1h | 9.80813 | 3.4716 | 1.54541 | 0.618519 |
| Chitin 1h | 7.82752 | 3.2797 | 1.3774 | 0.603478 |
| Epi 1h | 5.53738 | 3.21223 | 1.0248 | 0.593536 |
| SA 1h | 6.76537 | 3.26201 | 1.03156 | 0.523991 |
| Me-JA 1h | 11.0406 | 3.41195 | 1.9514 | 0.828213 |
| Control 6h | 15.1788 | 9.3094 | 1.68398 | 1.20565 |
| ABA 6h | 6.0095 | 3.48934 | 0.905839 | 0.525346 |
| ACC 6h | 7.01621 | 4.1631 | 0.95599 | 0.552598 |
| BABA 6h | 6.39991 | 3.71384 | 0.915357 | 0.530065 |
| Chitin 6h | 6.52133 | 3.77995 | 0.982586 | 0.56933 |
| Epi 6h | 9.0651 | 3.97519 | 1.2566 | 0.578647 |
| SA 6h | 9.19548 | 3.57512 | 1.35701 | 0.642373 |
| Me-JA 6h | 5.79972 | 3.36268 | 0.933209 | 0.540385 |
Source Transcript PGSC0003DMT400069918 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G39890.1 | +3 | 4e-79 | 266 | 147/216 (68%) | proline transporter 1 | chr2:16656022-16658202 FORWARD LENGTH=442 |
