Probe CUST_26633_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26633_PI426222305 | JHI_St_60k_v1 | DMT400000780 | GAAGTGCCAGACATAAATGCATTTGGTCAATCATTTAGCATAAACAGAATCTCATTTGCT |
All Microarray Probes Designed to Gene DMG401000287
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26523_PI426222305 | JHI_St_60k_v1 | DMT400000779 | GAAACTACTCTTCCTTCTGCTGCTTTATTTGTTATACGTTACCACTCTTTCTATGGTAAG |
| CUST_26579_PI426222305 | JHI_St_60k_v1 | DMT400000778 | TTTCCTCTTGGTTGTGCTTTTGATGAATCAATTGTTCACCACAAGGTATACTACTAATAC |
| CUST_26633_PI426222305 | JHI_St_60k_v1 | DMT400000780 | GAAGTGCCAGACATAAATGCATTTGGTCAATCATTTAGCATAAACAGAATCTCATTTGCT |
Microarray Signals from CUST_26633_PI426222305
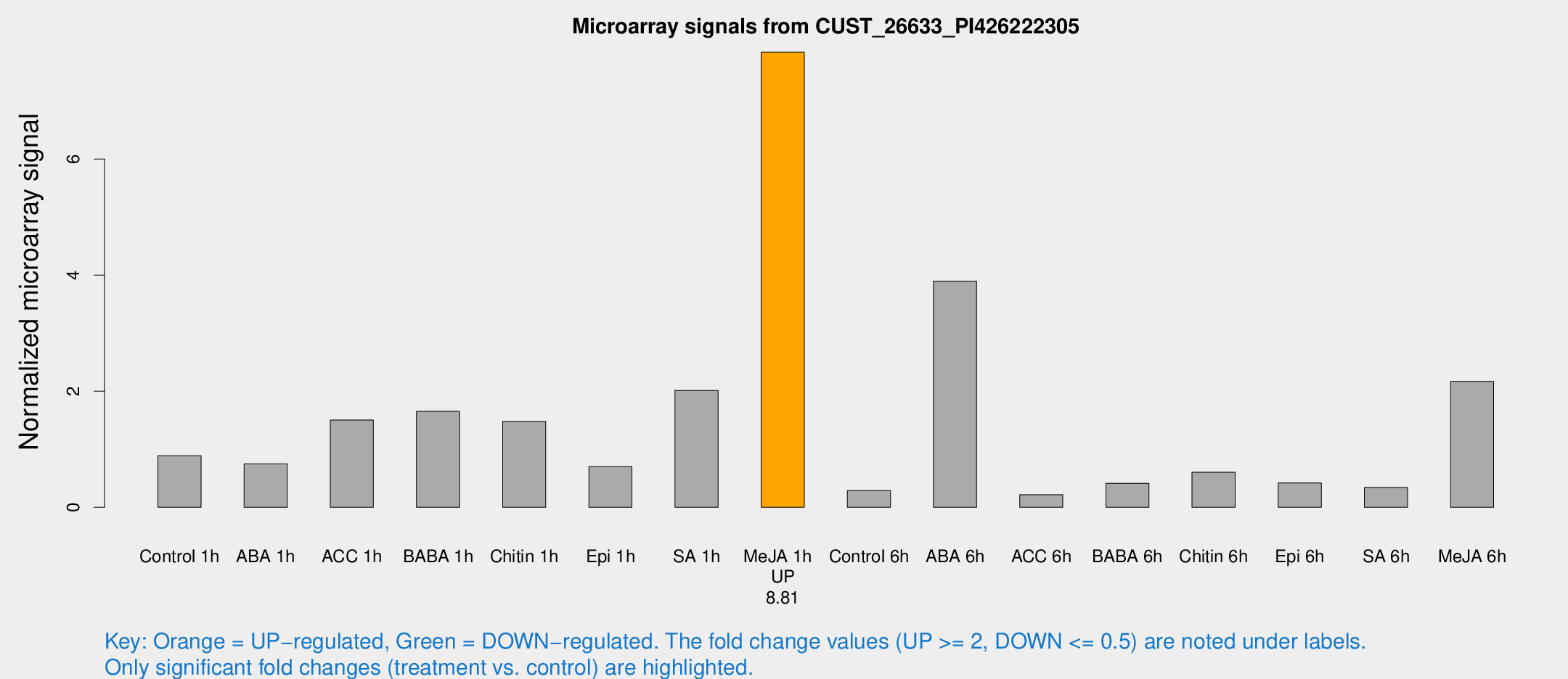
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 27.857 | 3.89018 | 0.889953 | 0.119073 |
| ABA 1h | 20.6538 | 3.25563 | 0.748607 | 0.121775 |
| ACC 1h | 49.7649 | 11.4665 | 1.50304 | 0.332408 |
| BABA 1h | 50.23 | 7.64908 | 1.6534 | 0.155208 |
| Chitin 1h | 45.011 | 14.0663 | 1.47841 | 0.454642 |
| Epi 1h | 21.7053 | 6.98515 | 0.699838 | 0.333686 |
| SA 1h | 73.1862 | 28.2759 | 2.01336 | 0.787805 |
| Me-JA 1h | 207.46 | 47.0794 | 7.84098 | 1.1882 |
| Control 6h | 10.7638 | 4.9909 | 0.287265 | 0.132704 |
| ABA 6h | 168.924 | 82.1016 | 3.89818 | 3.00012 |
| ACC 6h | 7.81351 | 4.07344 | 0.217383 | 0.112778 |
| BABA 6h | 16.9518 | 6.16801 | 0.414451 | 0.191436 |
| Chitin 6h | 54.1978 | 46.9838 | 0.603614 | 1.58217 |
| Epi 6h | 17.261 | 7.04514 | 0.420264 | 0.179067 |
| SA 6h | 10.9758 | 3.54862 | 0.340429 | 0.129285 |
| Me-JA 6h | 70.3463 | 17.813 | 2.17074 | 0.565375 |
Source Transcript PGSC0003DMT400000780 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G26260.2 | +1 | 4e-169 | 479 | 234/318 (74%) | myo-inositol oxygenase 4 | chr4:13297939-13300146 FORWARD LENGTH=318 |
