Probe CUST_26579_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26579_PI426222305 | JHI_St_60k_v1 | DMT400000778 | TTTCCTCTTGGTTGTGCTTTTGATGAATCAATTGTTCACCACAAGGTATACTACTAATAC |
All Microarray Probes Designed to Gene DMG401000287
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26523_PI426222305 | JHI_St_60k_v1 | DMT400000779 | GAAACTACTCTTCCTTCTGCTGCTTTATTTGTTATACGTTACCACTCTTTCTATGGTAAG |
| CUST_26579_PI426222305 | JHI_St_60k_v1 | DMT400000778 | TTTCCTCTTGGTTGTGCTTTTGATGAATCAATTGTTCACCACAAGGTATACTACTAATAC |
| CUST_26633_PI426222305 | JHI_St_60k_v1 | DMT400000780 | GAAGTGCCAGACATAAATGCATTTGGTCAATCATTTAGCATAAACAGAATCTCATTTGCT |
Microarray Signals from CUST_26579_PI426222305
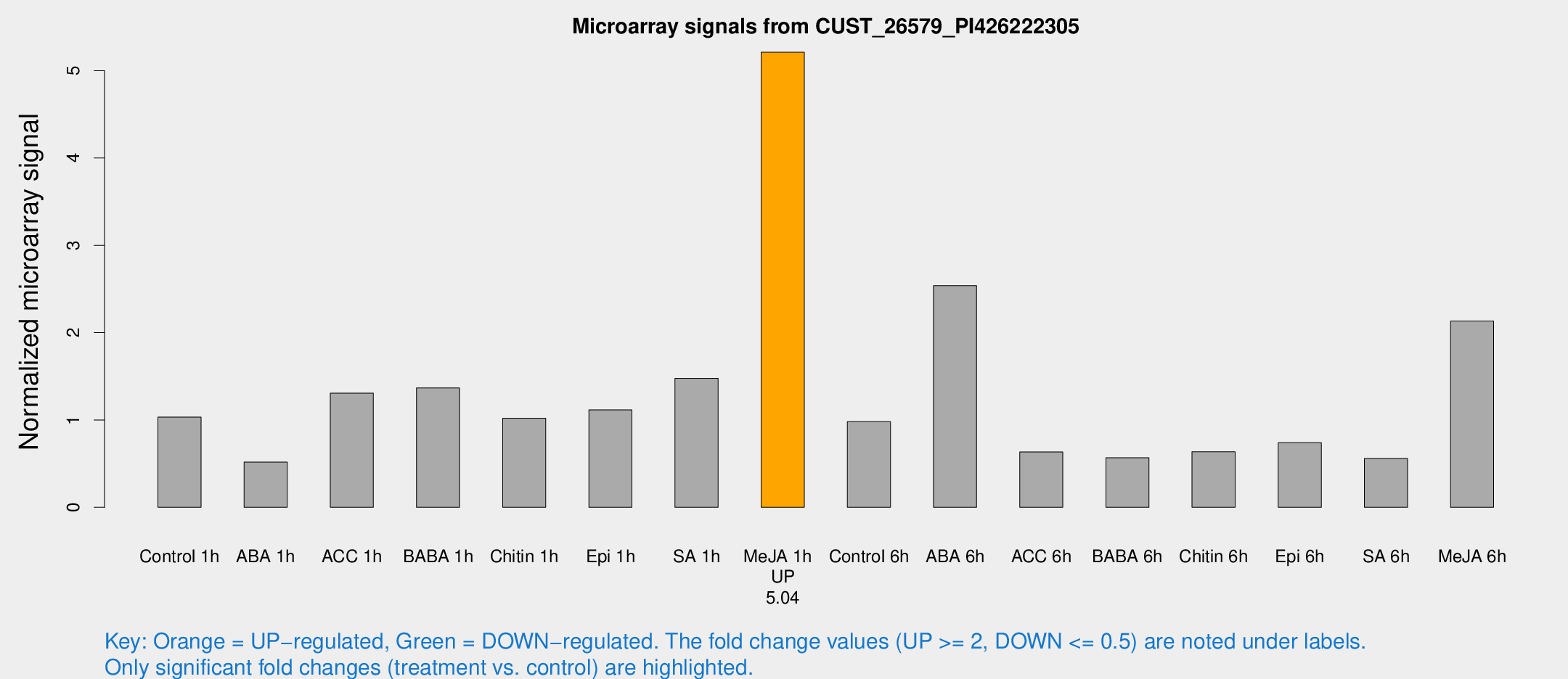
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 152.669 | 34.505 | 1.0336 | 0.226904 |
| ABA 1h | 69.2091 | 20.3816 | 0.516819 | 0.111336 |
| ACC 1h | 203.146 | 59.2709 | 1.30718 | 0.323864 |
| BABA 1h | 188.144 | 27.1402 | 1.36669 | 0.124242 |
| Chitin 1h | 132.347 | 22.2862 | 1.02035 | 0.147971 |
| Epi 1h | 135.894 | 9.70876 | 1.11577 | 0.0695064 |
| SA 1h | 251.862 | 103.712 | 1.47671 | 0.753741 |
| Me-JA 1h | 630.152 | 142.454 | 5.21091 | 0.843811 |
| Control 6h | 170.414 | 73.7641 | 0.980878 | 0.428795 |
| ABA 6h | 476.461 | 206.381 | 2.53869 | 1.69239 |
| ACC 6h | 108.645 | 28.1623 | 0.633683 | 0.0803848 |
| BABA 6h | 94.7849 | 21.4567 | 0.567562 | 0.133477 |
| Chitin 6h | 99.0927 | 20.3844 | 0.635417 | 0.158752 |
| Epi 6h | 155.584 | 65.3573 | 0.740494 | 0.691688 |
| SA 6h | 79.0011 | 10.863 | 0.558675 | 0.0413162 |
| Me-JA 6h | 305.568 | 52.368 | 2.13342 | 0.35631 |
Source Transcript PGSC0003DMT400000778 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G26260.2 | +1 | 1e-85 | 261 | 137/192 (71%) | myo-inositol oxygenase 4 | chr4:13297939-13300146 FORWARD LENGTH=318 |
