Probe CUST_26523_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26523_PI426222305 | JHI_St_60k_v1 | DMT400000779 | GAAACTACTCTTCCTTCTGCTGCTTTATTTGTTATACGTTACCACTCTTTCTATGGTAAG |
All Microarray Probes Designed to Gene DMG401000287
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26523_PI426222305 | JHI_St_60k_v1 | DMT400000779 | GAAACTACTCTTCCTTCTGCTGCTTTATTTGTTATACGTTACCACTCTTTCTATGGTAAG |
| CUST_26579_PI426222305 | JHI_St_60k_v1 | DMT400000778 | TTTCCTCTTGGTTGTGCTTTTGATGAATCAATTGTTCACCACAAGGTATACTACTAATAC |
| CUST_26633_PI426222305 | JHI_St_60k_v1 | DMT400000780 | GAAGTGCCAGACATAAATGCATTTGGTCAATCATTTAGCATAAACAGAATCTCATTTGCT |
Microarray Signals from CUST_26523_PI426222305
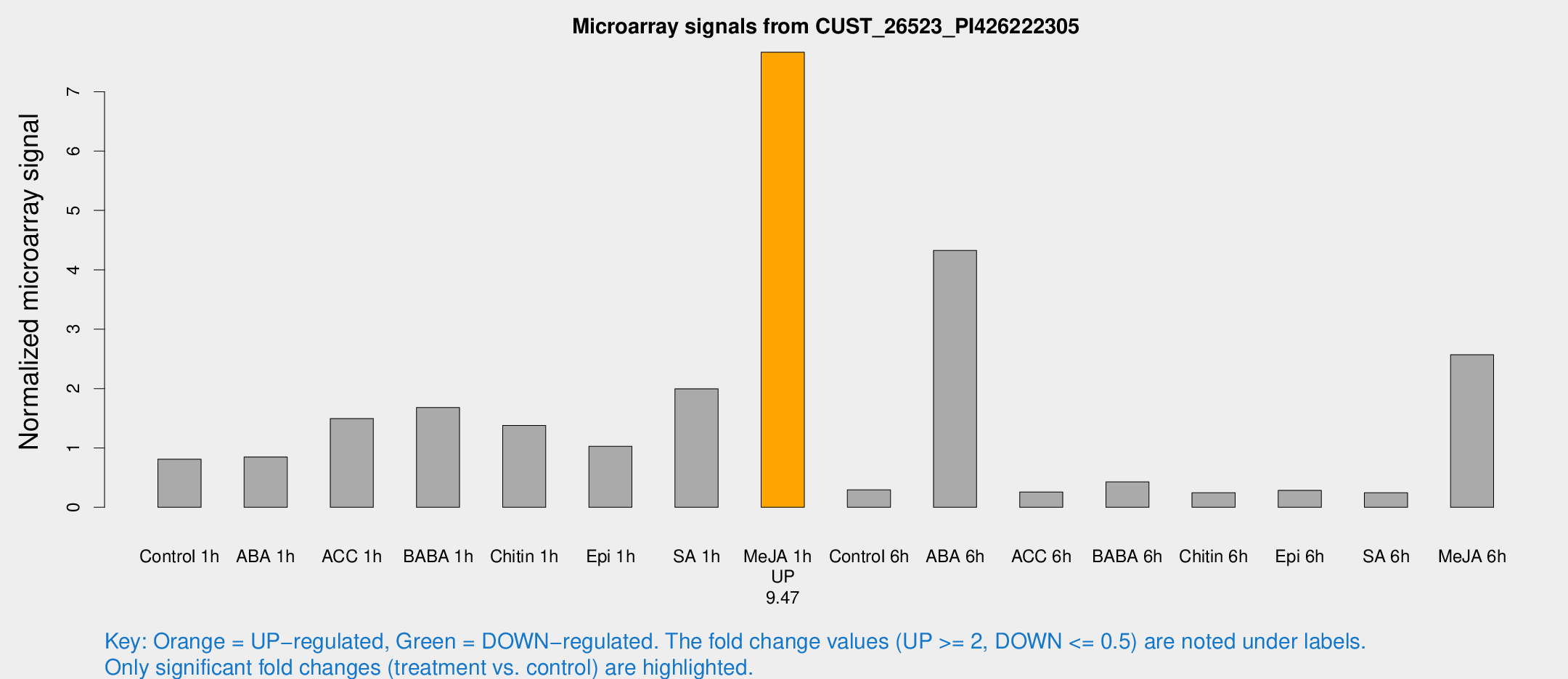
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 94.1059 | 19.1006 | 0.809594 | 0.202498 |
| ABA 1h | 91.1963 | 29.44 | 0.848184 | 0.217465 |
| ACC 1h | 176.42 | 35.4902 | 1.49439 | 0.323253 |
| BABA 1h | 182.382 | 24.0687 | 1.67978 | 0.102238 |
| Chitin 1h | 150.165 | 47.0534 | 1.38004 | 0.423332 |
| Epi 1h | 98.3581 | 6.49405 | 1.02775 | 0.0679035 |
| SA 1h | 267.537 | 114.087 | 1.99663 | 0.902165 |
| Me-JA 1h | 737.245 | 168.113 | 7.66454 | 1.46467 |
| Control 6h | 51.3811 | 30.1564 | 0.294518 | 0.260465 |
| ABA 6h | 725.161 | 419.802 | 4.32743 | 3.79336 |
| ACC 6h | 36.83 | 12.4269 | 0.258471 | 0.066088 |
| BABA 6h | 58.1836 | 17.9209 | 0.429126 | 0.11817 |
| Chitin 6h | 38.2912 | 15.4788 | 0.244723 | 0.215842 |
| Epi 6h | 58.926 | 32.2243 | 0.285786 | 0.478003 |
| SA 6h | 29.0594 | 8.3083 | 0.245611 | 0.0732309 |
| Me-JA 6h | 300.222 | 75.2071 | 2.5696 | 0.734628 |
Source Transcript PGSC0003DMT400000779 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G19800.1 | +1 | 1e-127 | 370 | 192/269 (71%) | myo-inositol oxygenase 2 | chr2:8531106-8533354 REVERSE LENGTH=317 |
