Probe CUST_25742_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25742_PI426222305 | JHI_St_60k_v1 | DMT400061790 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
All Microarray Probes Designed to Gene DMG402024041
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25835_PI426222305 | JHI_St_60k_v1 | DMT400061787 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25742_PI426222305 | JHI_St_60k_v1 | DMT400061790 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25951_PI426222305 | JHI_St_60k_v1 | DMT400061789 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25987_PI426222305 | JHI_St_60k_v1 | DMT400061791 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
Microarray Signals from CUST_25742_PI426222305
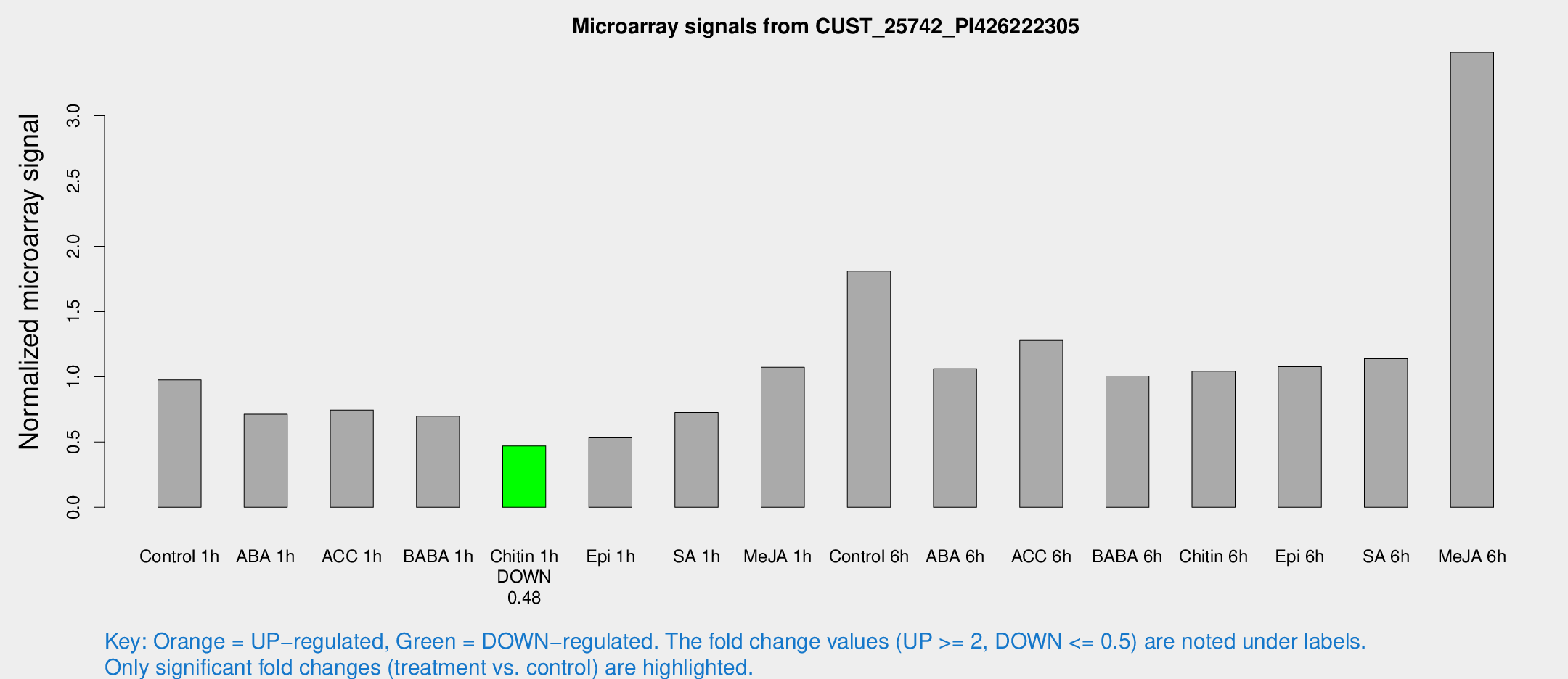
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 102.047 | 11.1409 | 0.976441 | 0.0630568 |
| ABA 1h | 67.8319 | 13.1286 | 0.712972 | 0.097163 |
| ACC 1h | 91.7673 | 28.8934 | 0.745412 | 0.279764 |
| BABA 1h | 70.0226 | 6.05977 | 0.697483 | 0.130153 |
| Chitin 1h | 43.9761 | 3.84166 | 0.47047 | 0.0410017 |
| Epi 1h | 48.8708 | 7.48426 | 0.532833 | 0.083668 |
| SA 1h | 87.9083 | 32.6791 | 0.726516 | 0.303897 |
| Me-JA 1h | 90.6572 | 6.02054 | 1.07263 | 0.0710579 |
| Control 6h | 193.967 | 34.8548 | 1.80926 | 0.177931 |
| ABA 6h | 118.666 | 14.829 | 1.06237 | 0.129713 |
| ACC 6h | 158.536 | 32.8605 | 1.27847 | 0.358223 |
| BABA 6h | 117.54 | 11.5516 | 1.00567 | 0.111685 |
| Chitin 6h | 117.96 | 20.4807 | 1.04219 | 0.190094 |
| Epi 6h | 134.108 | 30.5342 | 1.07763 | 0.394517 |
| SA 6h | 120.272 | 22.0566 | 1.13901 | 0.22889 |
| Me-JA 6h | 361.499 | 31.5389 | 3.48666 | 0.290773 |
Source Transcript PGSC0003DMT400061790 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G06350.1 | +3 | 3e-125 | 389 | 237/517 (46%) | dehydroquinate dehydratase, putative / shikimate dehydrogenase, putative | chr3:1924536-1927701 REVERSE LENGTH=603 |
