Probe CUST_25835_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25835_PI426222305 | JHI_St_60k_v1 | DMT400061787 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
All Microarray Probes Designed to Gene DMG402024041
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25835_PI426222305 | JHI_St_60k_v1 | DMT400061787 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25742_PI426222305 | JHI_St_60k_v1 | DMT400061790 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25951_PI426222305 | JHI_St_60k_v1 | DMT400061789 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
| CUST_25987_PI426222305 | JHI_St_60k_v1 | DMT400061791 | GGCCTCTTATTGGAACATGATTCATATATACAGTTCCCTTTCCCCTTGATAGTTAATATA |
Microarray Signals from CUST_25835_PI426222305
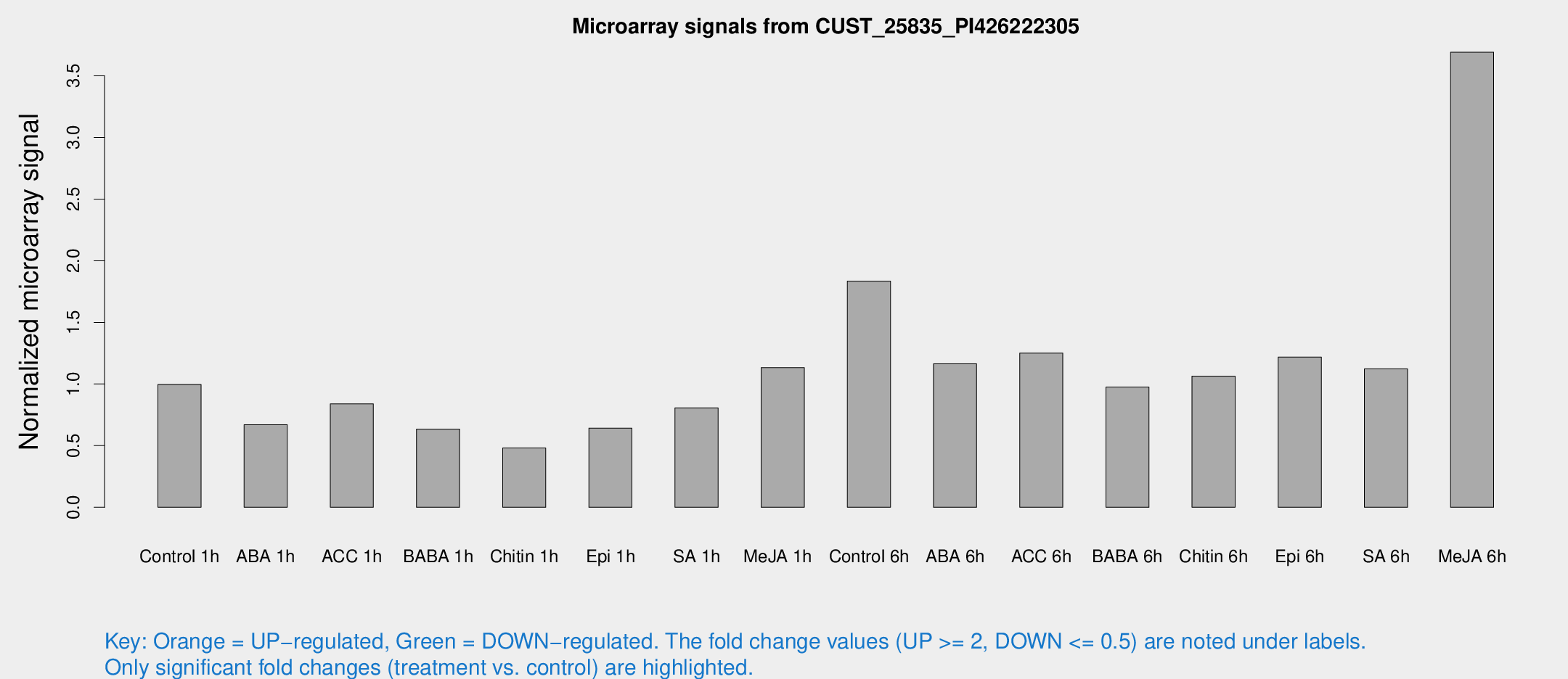
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 94.3813 | 10.8975 | 0.995564 | 0.103957 |
| ABA 1h | 57.3726 | 10.8632 | 0.670021 | 0.0774174 |
| ACC 1h | 88.0027 | 22.7695 | 0.839113 | 0.208642 |
| BABA 1h | 57.2466 | 4.62055 | 0.634598 | 0.0835621 |
| Chitin 1h | 40.6381 | 3.90359 | 0.480775 | 0.0459161 |
| Epi 1h | 52.6872 | 6.13465 | 0.641535 | 0.0739185 |
| SA 1h | 90.6492 | 37.1787 | 0.806146 | 0.369024 |
| Me-JA 1h | 89.3515 | 16.4887 | 1.13295 | 0.153696 |
| Control 6h | 186.553 | 52.5805 | 1.83487 | 0.381145 |
| ABA 6h | 116.705 | 10.7139 | 1.16389 | 0.08799 |
| ACC 6h | 138.227 | 22.727 | 1.2514 | 0.284321 |
| BABA 6h | 102.499 | 6.95427 | 0.9752 | 0.0986686 |
| Chitin 6h | 106.372 | 7.11868 | 1.06394 | 0.0880787 |
| Epi 6h | 136.315 | 29.8461 | 1.21795 | 0.428376 |
| SA 6h | 106.629 | 17.6415 | 1.12329 | 0.19019 |
| Me-JA 6h | 345.79 | 27.2202 | 3.6909 | 0.215933 |
Source Transcript PGSC0003DMT400061787 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G06350.1 | +1 | 1e-35 | 137 | 72/126 (57%) | dehydroquinate dehydratase, putative / shikimate dehydrogenase, putative | chr3:1924536-1927701 REVERSE LENGTH=603 |
